6YJ2
 
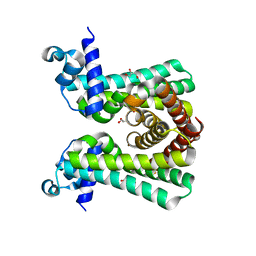 | | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism | | Descriptor: | GLYCEROL, Probable transcriptional regulatory protein (Probably TetR-family) | | Authors: | Keep, N.H, Pritchard, J.E, Sula, A, Cole, A.R, Kendall, S.L. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism
To be published
|
|
6YL2
 
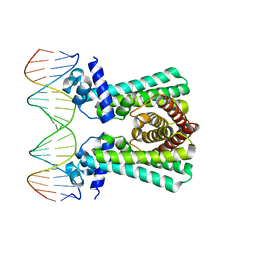 | | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*TP*AP*AP*TP*GP*AP*CP*GP*AP*TP*TP*AP*AP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*TP*TP*AP*AP*TP*CP*GP*TP*CP*AP*TP*TP*AP*AP*CP*GP*T)-3'), Probable transcriptional regulatory protein (Probably TetR-family) | | Authors: | Keep, N.H, Pritchard, J.E, Sula, A, Cole, A.R, Kendall, S.L. | | Deposit date: | 2020-04-06 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism
To be published
|
|
3MNL
 
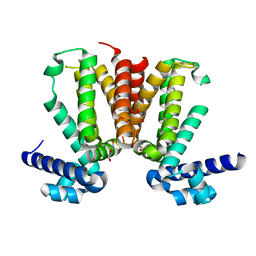 | | The crystal structure of KstR (Rv3574) from Mycobacterium tuberculosis H37Rv | | Descriptor: | TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY TETR-FAMILY), TRIETHYLENE GLYCOL | | Authors: | Gao, C, Bunker, R.D, ten Bokum, A, Kendall, S.L, Stoker, N.G, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-21 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J. Biol. Chem., 291, 2016
|
|
