4UR4
 
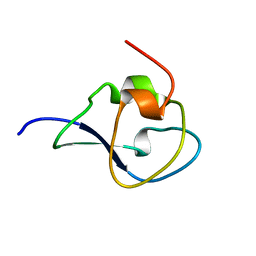 | | Structure of the type III fish antifreeze protein from Zoarces viviparus ZvAFP13 | | Descriptor: | ANTIFREEZE PROTEIN 13 | | Authors: | Wilkens, C, Poulsen, J.-C.N, Ramloev, H, Lo Leggio, L. | | Deposit date: | 2014-06-26 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Purification, Crystal Structure Determination and Functional Characterization of Type III Antifreeze Proteins from the European Eelpout Zoarces Viviparus.
Cryobiology, 69, 2014
|
|
6E98
 
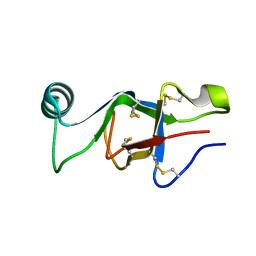 | |
6E9M
 
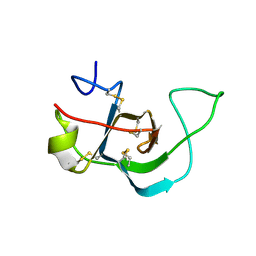 | |
6YWF
 
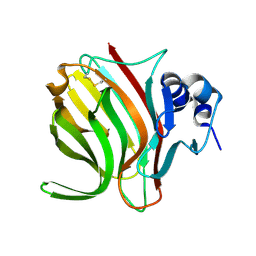 | |
8BJO
 
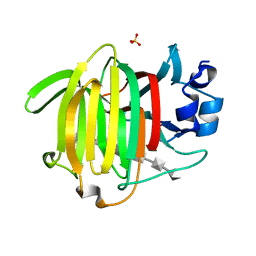 | |
7TB6
 
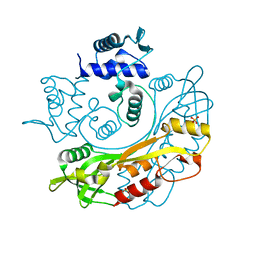 | | Structure of S. maltophilia CapW | | Descriptor: | S. maltophilia CapW, SULFATE ION | | Authors: | Blankenchip, C.L, Nguyen, J.V, Lau, R.K, Ye, Q, Corbett, K.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Control of bacterial immune signaling by a WYL domain transcription factor.
Nucleic Acids Res., 50, 2022
|
|
7TB5
 
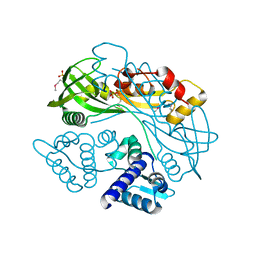 | | Structure of P. aeruginosa PA17 CapW | | Descriptor: | SULFATE ION, WYL domain-containing protein | | Authors: | Blankenchip, C.L, Nguyen, J.V, Lau, R.K, Ye, Q, Corbett, K.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Control of bacterial immune signaling by a WYL domain transcription factor.
Nucleic Acids Res., 50, 2022
|
|
8P6O
 
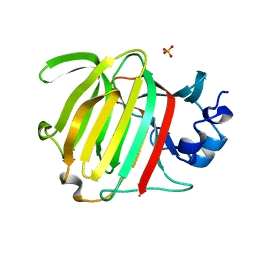 | |
7PXQ
 
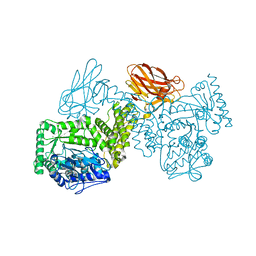 | | GH115 alpha-1,2-glucuronidase D303A | | Descriptor: | CALCIUM ION, xylan alpha-1,2-glucuronidase | | Authors: | Wilkens, C, Morth, J.P, Polikarpov, I. | | Deposit date: | 2021-10-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A GH115 alpha-glucuronidase structure reveals dimerization-mediated substrate binding and a proton wire potentially important for catalysis.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PUG
 
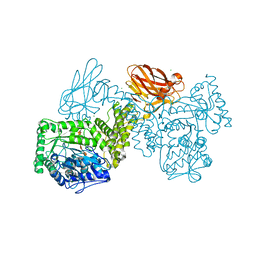 | | GH115 alpha-1,2-glucuronidase in complex with xylopentaose | | Descriptor: | CALCIUM ION, CHLORIDE ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Wilkens, C, Morth, J.P, Polikarpov, I. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A GH115 alpha-glucuronidase structure reveals dimerization-mediated substrate binding and a proton wire potentially important for catalysis.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
2WGO
 
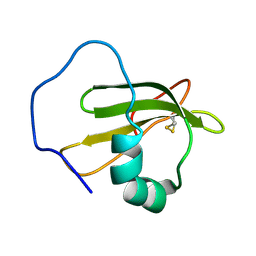 | | Structure of ranaspumin-2, a surfactant protein from the foam nests of a tropical frog | | Descriptor: | RANASPUMIN-2 | | Authors: | Mackenzie, C.D, Smith, B.O, Kennedy, M.W, Cooper, A. | | Deposit date: | 2009-04-21 | | Release date: | 2009-06-23 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Ranaspumin-2: Structure and Function of a Surfactant Protein from the Foam Nests of a Tropical Frog.
Biophys.J., 96, 2009
|
|
4UR6
 
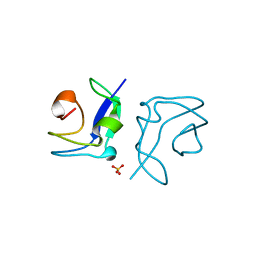 | | Structure of the type III fish antifreeze protein from Zoarces viviparus ZvAFP6 | | Descriptor: | SULFATE ION, TYPE III ANTIFREEZE PROTEIN 6 | | Authors: | Wilkens, C, Poulsen, J.-C.N, Ramloev, H, Lo Leggio, L. | | Deposit date: | 2014-06-26 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Purification, Crystal Structure Determination and Functional Characterization of Type III Antifreeze Proteins from the European Eelpout Zoarces Viviparus.
Cryobiology, 69, 2014
|
|
8PED
 
 | |
7QFY
 
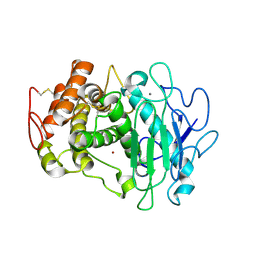 | | Fusarium oxysporum M36 protease without the propeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular metalloproteinase, ... | | Authors: | Wilkens, C, Qiu, J, Meyer, A.S, Morth, J.P. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fusarium oxysporum M36 protease without the propeptide
To Be Published
|
|
7QP3
 
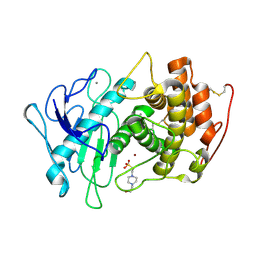 | | Pseudogymnoascus pannorum M36 protease without the propeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wilkens, C, Qiu, J, Meyer, A.S, Morth, J.P. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Phaeosphaeria nodorum M36 protease without the propeptide
To Be Published
|
|
2LVS
 
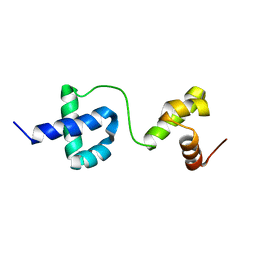 | |
8BXZ
 
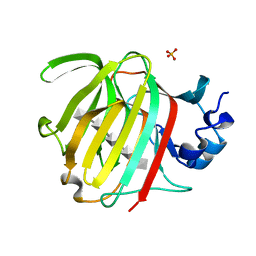 | |
8BZK
 
 | |
8C0M
 
 | |
5W0Y
 
 | |
2Y7C
 
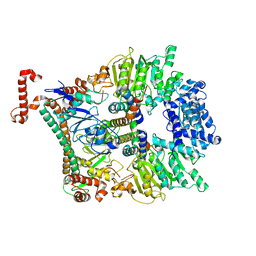 | | Atomic model of the Ocr-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | GENE 0.3 PROTEIN, TYPE I RESTRICTION ENZYME ECOKI M PROTEIN, TYPE-1 RESTRICTION ENZYME ECOKI SPECIFICITY PROTEIN | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
7Z6T
 
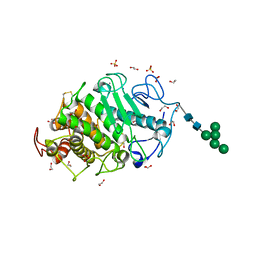 | | Aspergillus clavatus M36 protease without the propeptide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Extracellular metalloproteinase mep, ... | | Authors: | Wilkens, C, Qiu, J, Meyer, A.S, Morth, J.P. | | Deposit date: | 2022-03-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Aspergillus clavatus M36 protease without the propeptide
To Be Published
|
|
8BF3
 
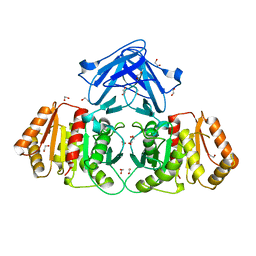 | |
2Y7H
 
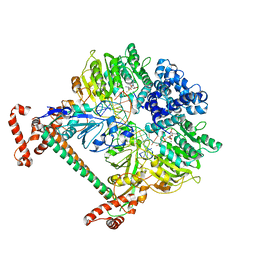 | | Atomic model of the DNA-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | 5'-D(*GP*TP*TP*CP*AP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*GP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*CP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*TP*GP*AP*AP*C)-3', S-ADENOSYLMETHIONINE, ... | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
8BBP
 
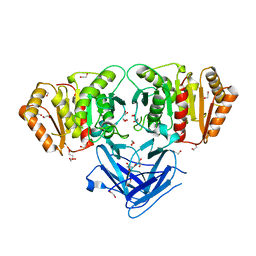 | |
