7GTF
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with XST00000754b | | Descriptor: | 1,2-benzoxazol-3-ol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTP
 
 | |
7GTV
 
 | |
7GTR
 
 | |
7GTY
 
 | |
7GU5
 
 | |
7GU9
 
 | |
7GUB
 
 | |
7GU3
 
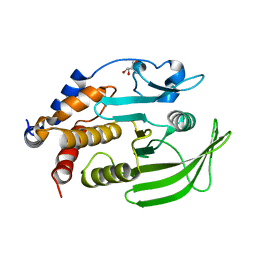 | |
7GU6
 
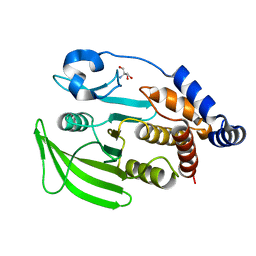 | |
7GU1
 
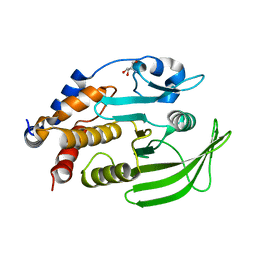 | |
7GTW
 
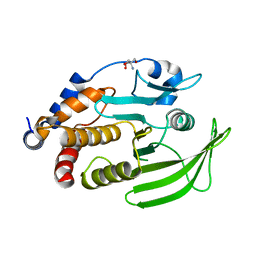 | |
7GU2
 
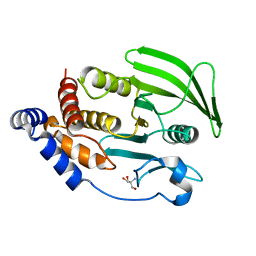 | |
7GTZ
 
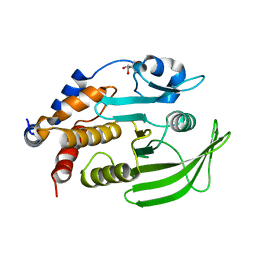 | |
7GU4
 
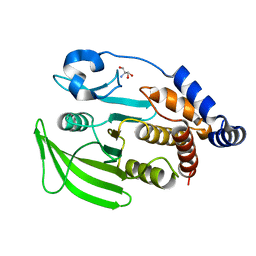 | |
7GTX
 
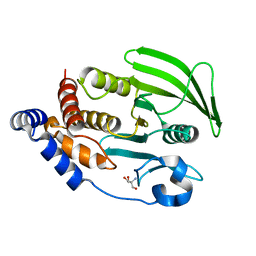 | |
7GU0
 
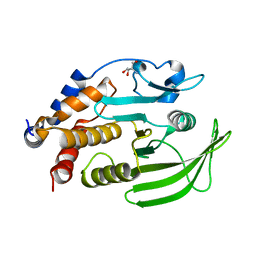 | |
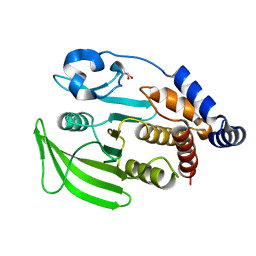
7GUA
 
 | |
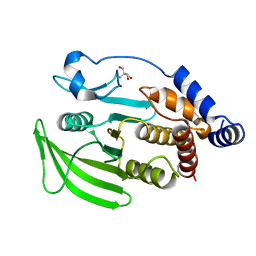
7GU7
 
 | |
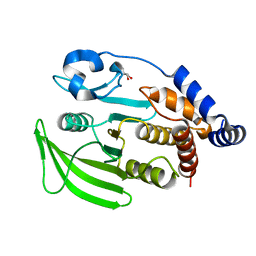
7GU8
 
 | |
7GUC
 
 | |
7MHI
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 298 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHQ
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 310 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9601 Å) | | Cite: | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MHN
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 277 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1908 Å) | | Cite: | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MHJ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 298 K and High Humidity | | Descriptor: | 3C-like proteinase, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.0005 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
