4GIO
 
 | | Crystal structure of Campylobacter jejuni cj0090 | | 分子名称: | BROMIDE ION, Putative lipoprotein | | 著者 | Kawai, F, Yeo, H.J. | | 登録日 | 2012-08-08 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the Campylobacter jejuni Cj0090 protein reveals a novel variant of the immunoglobulin fold among bacterial lipoproteins.
Proteins, 80, 2012
|
|
3Q3I
 
 | |
3Q3H
 
 | |
3Q3E
 
 | |
5ZL9
 
 | |
3UAU
 
 | |
3A3F
 
 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae,complexed with novel beta-lactam (FMZ) | | 分子名称: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxo-1-({(2R)-2-[(2-oxoimidazolidin-1-yl)amino]-2-phenylacetyl}amino)ethyl]-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4 | | 著者 | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2009-06-12 | | 公開日 | 2009-12-22 | | 最終更新日 | 2013-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
3A3I
 
 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae, complexed with ampicillin (AIX) | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4 | | 著者 | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2009-06-12 | | 公開日 | 2009-12-22 | | 最終更新日 | 2013-11-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
3A3D
 
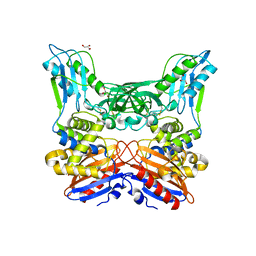 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae | | 分子名称: | GLYCEROL, Penicillin-binding protein 4 | | 著者 | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2009-06-12 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
3A3E
 
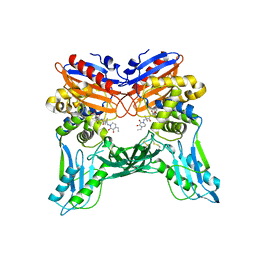 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae, complexed with novel beta-lactam (CMV) | | 分子名称: | (2R,4S)-2-[(1R)-1-({(2R)-2-[(4-ethyl-2,3-dioxopiperazin-1-yl)amino]-2-phenylacetyl}amino)-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4 | | 著者 | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2009-06-12 | | 公開日 | 2009-12-22 | | 最終更新日 | 2013-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
3A3J
 
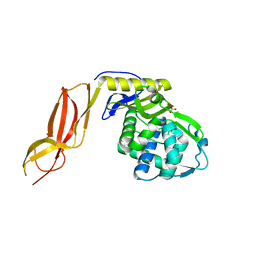 | | Crystal structures of penicillin binding protein 5 from Haemophilus influenzae | | 分子名称: | PBP5, SULFATE ION | | 著者 | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2009-06-12 | | 公開日 | 2009-12-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
2NOO
 
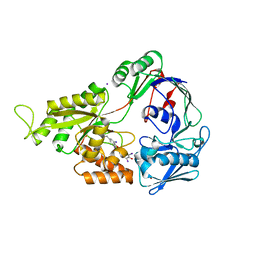 | | Crystal Structure of Mutant NikA | | 分子名称: | IODIDE ION, NICKEL (II) ION, Nickel-binding periplasmic protein | | 著者 | Addy, C, Ohara, M, Kawai, F, Kidera, A, Ikeguchi, M, Fuchigami, S, Osawa, M, Shimada, I, Park, S.Y, Tame, J.R.H, Heddle, J.G. | | 登録日 | 2006-10-26 | | 公開日 | 2007-01-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Nickel binding to NikA: an additional binding site reconciles spectroscopy, calorimetry and crystallography.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4YUS
 
 | | Crystal structure of photoactivated adenylyl cyclase of a cyanobacteriaOscillatoria acuminata in hexagonal form | | 分子名称: | FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase | | 著者 | Park, S.-Y, Ohki, M, Sugiyama, K, Kawai, F, Iseki, M. | | 登録日 | 2015-03-19 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insight into photoactivation of an adenylate cyclase from a photosynthetic cyanobacterium
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4WFJ
 
 | | Crystal structure of PET-degrading cutinase Cut190 S226P mutant in Ca(2+)-bound state at 1.75 angstrom resolution | | 分子名称: | CALCIUM ION, CHLORIDE ION, Cutinase | | 著者 | Miyakawa, T, Mizushima, H, Ohtsuka, J, Oda, M, Kawai, F, Tanokura, M. | | 登録日 | 2014-09-15 | | 公開日 | 2014-12-24 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4WFK
 
 | | Crystal structure of PET-degrading cutinase Cut190 S226P mutant in Ca(2+)-bound state at 2.35 angstrom resolution | | 分子名称: | CALCIUM ION, CHLORIDE ION, Cutinase | | 著者 | Miyakawa, T, Mizushima, H, Ohtsuka, J, Oda, M, Kawai, F, Tanokura, M. | | 登録日 | 2014-09-15 | | 公開日 | 2014-12-24 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4WFI
 
 | | Crystal structure of PET-degrading cutinase Cut190 S226P mutant in Ca(2+)-free state | | 分子名称: | Cutinase | | 著者 | Miyakawa, T, Mizushima, H, Ohtsuka, J, Oda, M, Kawai, F, Tanokura, M. | | 登録日 | 2014-09-15 | | 公開日 | 2014-12-24 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.446 Å) | | 主引用文献 | Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
7ECD
 
 | | Crystal structure of Tam41 from Firmicutes bacterium, complex with CTP-Mg | | 分子名称: | BROMIDE ION, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Kimura, K, Kawai, F, Kubota-Kawai, H, Watanabe, Y, Tamura, Y. | | 登録日 | 2021-03-12 | | 公開日 | 2022-01-19 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of Tam41 cytidine diphosphate diacylglycerol synthase from a Firmicutes bacterium.
J.Biochem., 171, 2022
|
|
7CTR
 
 | | Closed form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, Alpha/beta hydrolase family protein | | 著者 | Emori, M, Numoto, N, Senga, A, Bekker, G.J, Kamiya, N, Ito, N, Kawai, F, Oda, M. | | 登録日 | 2020-08-20 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural basis of mutants of PET-degrading enzyme from Saccharomonospora viridis AHK190 with high activity and thermal stability.
Proteins, 89, 2021
|
|
7CEF
 
 | | Crystal structure of PET-degrading cutinase Cut190 /S226P/R228S/ mutant with the C-terminal three residues deletion | | 分子名称: | Alpha/beta hydrolase family protein, CALCIUM ION, ZINC ION | | 著者 | Senga, A, Numoto, N, Ito, N, Kawai, F, Oda, M. | | 登録日 | 2020-06-23 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Multiple structural states of Ca2+-regulated PET hydrolase, Cut190, and its correlation with activity and stability.
J.Biochem., 169, 2021
|
|
7CEH
 
 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S/ mutant with the C-terminal three residues deletion in ligand ejecting form | | 分子名称: | Alpha/beta hydrolase family protein, CALCIUM ION | | 著者 | Senga, A, Numoto, N, Ito, N, Kawai, F, Oda, M. | | 登録日 | 2020-06-23 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Multiple structural states of Ca2+-regulated PET hydrolase, Cut190, and its correlation with activity and stability.
J.Biochem., 169, 2021
|
|
7CTS
 
 | | Open form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H and S176A inactivation | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, Alpha/beta hydrolase family protein, BICINE, ... | | 著者 | Emori, M, Numoto, N, Senga, A, Bekker, G.J, Kamiya, N, Ito, N, Kawai, F, Oda, M. | | 登録日 | 2020-08-20 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structural basis of mutants of PET-degrading enzyme from Saccharomonospora viridis AHK190 with high activity and thermal stability.
Proteins, 89, 2021
|
|
4YUT
 
 | | Crystal structure of photoactivated adenylyl cyclase of a cyanobacteriaOscillatoria acuminata in orthorhombic form | | 分子名称: | FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase | | 著者 | Park, S.-Y, Ohki, M, Sugiyama, K, Kawai, F, Iseki, M. | | 登録日 | 2015-03-19 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural insight into photoactivation of an adenylate cyclase from a photosynthetic cyanobacterium
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ZNO
 
 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S/ mutant in Ca(2+)-bound state | | 分子名称: | Alpha/beta hydrolase family protein, CALCIUM ION, GLYCEROL | | 著者 | Numoto, N, Inaba, S, Yamagami, Y, Kamiya, N, Bekker, G.J, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | 登録日 | 2018-04-10 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.60264349 Å) | | 主引用文献 | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
5ZRS
 
 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S mutant in monoethyl adipate bound state | | 分子名称: | 6-ethoxy-6-oxohexanoic acid, Alpha/beta hydrolase family protein, CALCIUM ION, ... | | 著者 | Numoto, N, Kamiya, N, Bekker, G.J, Yamagami, Y, Inaba, S, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | 登録日 | 2018-04-25 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
5ZRR
 
 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S mutant in monoethyl succinate bound state | | 分子名称: | 4-ethoxy-4-oxobutanoic acid, Alpha/beta hydrolase family protein, GLYCEROL, ... | | 著者 | Numoto, N, Kamiya, N, Bekker, G.J, Yamagami, Y, Inaba, S, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | 登録日 | 2018-04-25 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
