5BW7
 
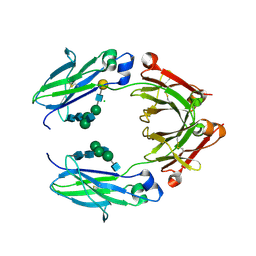 | | Crystal structure of nonfucosylated Fc Y296W mutant complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Isoda, Y, Yagi, H, Satoh, T, Shibata-Koyama, M, Masuda, K, Satoh, M, Kato, K, Iida, S. | | Deposit date: | 2015-06-06 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Importance of the Side Chain at Position 296 of Antibody Fc in Interactions with Fc gamma RIIIa and Other Fc gamma Receptors
Plos One, 10, 2015
|
|
4WLC
 
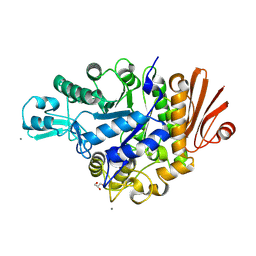 | | Structure of dextran glucosidase with glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Glucan 1,6-alpha-glucosidase, ... | | Authors: | Kobayashi, M, Kato, K, Yao, M. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural insights into the catalytic reaction that is involved in the reorientation of Trp238 at the substrate-binding site in GH13 dextran glucosidase
Febs Lett., 589, 2015
|
|
2RU6
 
 | | The pure alternative state of ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Kitazawa, S, Kameda, T, Kumo, A, Utsumi, M, Baxter, N, Kato, K, Williamson, M.P, Kitahara, R. | | Deposit date: | 2013-12-04 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Close Identity between Alternatively Folded State N2 of Ubiquitin and the Conformation of the Protein Bound to the Ubiquitin-Activating Enzyme
Biochemistry, 53, 2014
|
|
4XB3
 
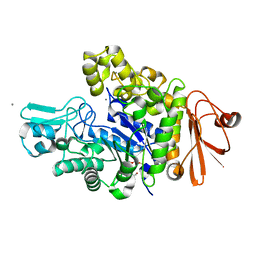 | | Structure of dextran glucosidase | | Descriptor: | CALCIUM ION, Glucan 1,6-alpha-glucosidase, HEXAETHYLENE GLYCOL | | Authors: | Kobayashi, M, Kato, K, Yao, M. | | Deposit date: | 2014-12-16 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Structural insights into the catalytic reaction that is involved in the reorientation of Trp238 at the substrate-binding site in GH13 dextran glucosidase
Febs Lett., 589, 2015
|
|
5DSV
 
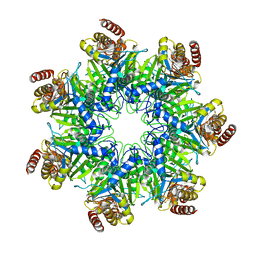 | | Crystal structure of human proteasome alpha7 tetradecamer | | Descriptor: | Proteasome subunit alpha type-3 | | Authors: | Satoh, T, Thammaporn, R, Seetaha, S, Kato, K. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Disassembly of the self-assembled, double-ring structure of proteasome alpha 7 homo-tetradecamer by alpha 6
Sci Rep, 5, 2015
|
|
6S22
 
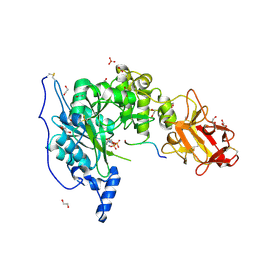 | | Crystal structure of the TgGalNAc-T3 in complex with UDP, manganese and FGF23c | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | de las Rivas, M, Daniel, E.J.P, Narimatsu, Y, Companon, I, Kato, K, Hermosilla, P, Thureau, A, Ceballos-Laita, L, Coelho, H, Bernado, P, Marcelo, F, Hansen, L, Lostao, A, Corzana, F, Clausen, H, Gerken, T.A, Hurtado-Guerrero, R. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Molecular basis for fibroblast growth factor 23 O-glycosylation by GalNAc-T3.
Nat.Chem.Biol., 16, 2020
|
|
6S24
 
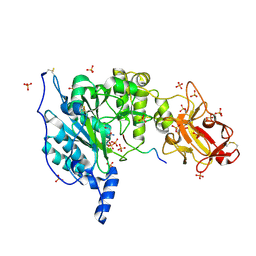 | | Crystal structure of the TgGalNAc-T3 in complex with UDP, manganese and the peptide 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALA-THR-GLY-ALA-GLY-ALA-GLY-ALA-GLY-THR-THR-PRO-GLY-PRO, ... | | Authors: | de las Rivas, M, Daniel, E.J.P, Narimatsu, Y, Companon, I, Kato, K, Hermosilla, P, Thureau, A, Ceballos-Laita, L, Coelho, H, Bernado, P, Marcelo, F, Hansen, L, Lostao, A, Corzana, F, Clausen, H, Gerken, T.A, Hurtado-Guerrero, R. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Molecular basis for fibroblast growth factor 23 O-glycosylation by GalNAc-T3.
Nat.Chem.Biol., 16, 2020
|
|
1UMI
 
 | | Structural basis of sugar-recognizing ubiquitin ligase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, F-box only protein 2 | | Authors: | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K. | | Deposit date: | 2003-10-01 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of sugar-recognizing ubiquitin ligase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UMH
 
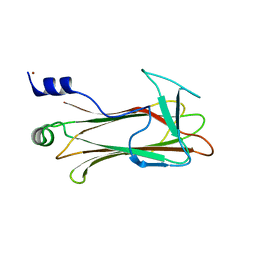 | | Structural basis of sugar-recognizing ubiquitin ligase | | Descriptor: | F-box only protein 2, NICKEL (II) ION | | Authors: | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-01 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of sugar-recognizing ubiquitin ligase
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
8JH0
 
 | | Crystal structure of the light-driven sodium pump IaNaR | | Descriptor: | RETINAL, Xanthorhodopsin | | Authors: | Hashimoto, T, Kato, K, Tanaka, Y, Yao, M, Kikukawa, T. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Multistep conformational changes leading to the gate opening of light-driven sodium pump rhodopsin.
J.Biol.Chem., 299, 2023
|
|
4YGE
 
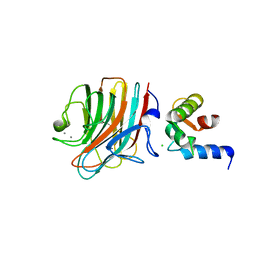 | | Crystal structure of ERGIC-53/MCFD2, trigonal calcium-bound form 2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Satoh, T, Nishio, M, Yagi-Utsumi, M, Suzuki, K, Anzai, T, Mizushima, T, Kamiya, Y, Kato, K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystallographic snapshots of the EF-hand protein MCFD2 complexed with the intracellular lectin ERGIC-53 involved in glycoprotein transport.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4YGB
 
 | | Crystal structure of ERGIC-53/MCFD2, monoclinic calcium-free form | | Descriptor: | CALCIUM ION, GLYCEROL, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Satoh, T, Nishio, M, Yagi-Utsumi, M, Suzuki, K, Anzai, T, Mizushima, T, Kamiya, Y, Kato, K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic snapshots of the EF-hand protein MCFD2 complexed with the intracellular lectin ERGIC-53 involved in glycoprotein transport.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1WUA
 
 | | The structure of Aplyronine A-actin complex | | Descriptor: | (8R,9R,10R,11R,14S,18S,20S,24S)-24-{(1R,2S,3R,6R,7R,8R,9S,10E)-8-(ACETYLOXY)-6-[(N,N-DIMETHYLALANYL)OXY]-11-[FORMYL(MET HYL)AMINO]-2-HYDROXY-1,3,7,9-TETRAMETHYLUNDEC-10-ENYL}-10-HYDROXY-14,20-DIMETHOXY-9,11,15,18-TETRAMETHYL-2-OXOOXACYCLOTE TRACOSA-3,5,15,21-TETRAEN-8-YL N,N,O-TRIMETHYLSERINATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Hirata, K, Muraoka, S, Suenaga, K, Kuroda, T, Kato, K, Tanaka, H, Yamamoto, M, Takata, M, Yamada, K, Kigoshi, H. | | Deposit date: | 2004-12-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure basis for antitumor effect of aplyronine a
J.Mol.Biol., 356, 2006
|
|
4YGC
 
 | | Crystal structure of ERGIC-53/MCFD2, monoclinic calcium-bound form 1 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Satoh, T, Nishio, M, Yagi-Utsumi, M, Suzuki, K, Anzai, T, Mizushima, T, Kamiya, Y, Kato, K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic snapshots of the EF-hand protein MCFD2 complexed with the intracellular lectin ERGIC-53 involved in glycoprotein transport.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4YGD
 
 | | Crystal structure of ERGIC-53/MCFD2, monoclinic calcium-bound form 2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Satoh, T, Nishio, M, Yagi-Utsumi, M, Suzuki, K, Anzai, T, Mizushima, T, Kamiya, Y, Kato, K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystallographic snapshots of the EF-hand protein MCFD2 complexed with the intracellular lectin ERGIC-53 involved in glycoprotein transport.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1WXS
 
 | | Solution Structure of Ufm1, a ubiquitin-fold modifier | | Descriptor: | Ubiquitin-fold Modifier 1 | | Authors: | Sasakawa, H, Sakata, E, Yamaguchi, Y, Komatsu, M, Tatsumi, K, Kominami, E, Tanaka, K, Kato, K. | | Deposit date: | 2005-02-01 | | Release date: | 2006-04-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of Ufm1, a ubiquitin-fold modifier 1
Biochem.Biophys.Res.Commun., 343, 2006
|
|
8H3M
 
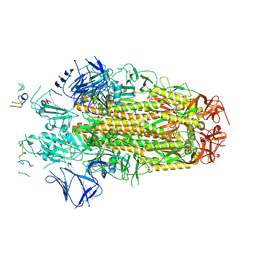 | | Conformation 1 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy chain, Spike glycoprotein | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
8H3N
 
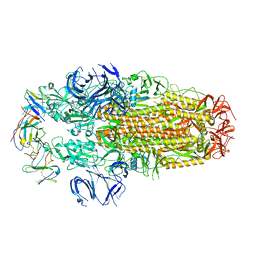 | | Conformation 2 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy-chain, MO1 light chain, ... | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
5AYE
 
 | | Crystal structure of Ruminococcus albus beta-(1,4)-mannooligosaccharide phosphorylase (RaMP2) in complexes with phosphate and beta-(1,4)-mannobiose | | Descriptor: | Beta-1,4-mannooligosaccharide phosphorylase, PHOSPHATE ION, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Ye, Y, Saburi, W, Kato, K, Yao, M. | | Deposit date: | 2015-08-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the difference in substrate recognition of two mannoside phosphorylases from two GH130 subfamilies.
Febs Lett., 590, 2016
|
|
5AY9
 
 | |
5AYD
 
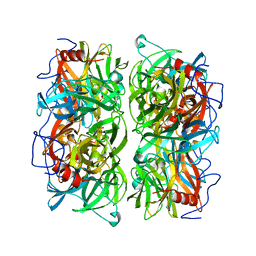 | | Crystal structure of Ruminococcus albus beta-(1,4)-mannooligosaccharide phosphorylase (RaMP2) in complexes with phosphate | | Descriptor: | Beta-1,4-mannooligosaccharide phosphorylase, PHOSPHATE ION | | Authors: | Ye, Y, Saburi, W, Kato, K, Yao, M. | | Deposit date: | 2015-08-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the difference in substrate recognition of two mannoside phosphorylases from two GH130 subfamilies.
Febs Lett., 590, 2016
|
|
5AYC
 
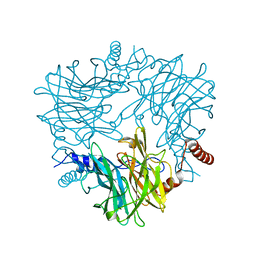 | | Crystal structure of Ruminococcus albus 4-O-beta-D-mannosyl-D-glucose phosphorylase (RaMP1) in complexes with sulfate and 4-O-beta-D-mannosyl-D-glucose | | Descriptor: | 4-O-beta-D-mannosyl-D-glucose phosphorylase, SULFATE ION, beta-D-mannopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Ye, Y, Saburi, W, Kato, K, Yao, M. | | Deposit date: | 2015-08-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the difference in substrate recognition of two mannoside phosphorylases from two GH130 subfamilies.
Febs Lett., 590, 2016
|
|
5H18
 
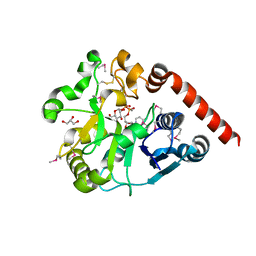 | | Crystal structure of catalytic domain of UGGT (UDP-glucose-bound form) from Thermomyces dupontii | | Descriptor: | CALCIUM ION, GLYCEROL, UGGT, ... | | Authors: | Satoh, T, Zhu, T, Toshimori, T, Kamikubo, H, Uchihashi, T, Kato, K. | | Deposit date: | 2016-10-08 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Visualisation of a flexible modular structure of the ER folding-sensor enzyme UGGT.
Sci Rep, 7, 2017
|
|
5DKZ
 
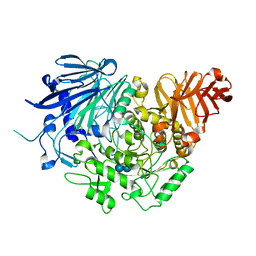 | | Crystal structure of glucosidase II alpha subunit (alpha3-Glc2-bound from) | | Descriptor: | Alpha glucosidase-like protein, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
5DKY
 
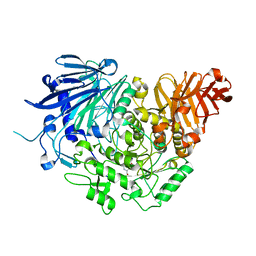 | | Crystal structure of glucosidase II alpha subunit (DNJ-bound from) | | Descriptor: | 1-DEOXYNOJIRIMYCIN, Alpha glucosidase-like protein | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
