1SMW
 
 | | Crystal Structure of Cp Rd L41A mutant in reduced state 2 (soaked) | | Descriptor: | FE (II) ION, Rubredoxin | | Authors: | Park, I.Y, Youn, B, Harley, J.L, Eidsness, M.K, Smith, E, Ichiye, T, Kang, C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The unique hydrogen bonded water in the reduced form of Clostridium pasteurianum rubredoxin and its possible role in electron transfer
J.BIOL.INORG.CHEM., 9, 2004
|
|
1T4I
 
 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*AP*AP*TP*TP*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.S, Kang, C. | | Deposit date: | 2004-04-29 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a DNA Decamer Containing a cis-syn Thymine-dimer
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1T9O
 
 | | Crystal Structure of V44G Cp Rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Park, I.Y, Eidsness, M.K, Lin, I.J, Gebel, E.B, Youn, B, Harley, J.L, Machonkin, T.E, Frederick, R.O, Markley, J.L, Smith, E.T, Ichiye, T, Kang, C. | | Deposit date: | 2004-05-18 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies of V44 mutants of Clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential.
Proteins, 57, 2004
|
|
1RF4
 
 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase, Tetrahedral intermediate Bound State | | Descriptor: | (3R,4S,5R)-5-{[(1R)-1-CARBOXY-2-FLUORO-1-(PHOSPHONOOXY)ETHYL]OXY}-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 5-enolpyruvylshikimate-3-phosphate synthase | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
4G0K
 
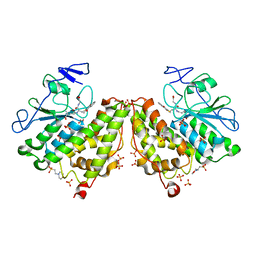 | | Glutathionyl-hydroquinone reductase, YqjG, of E.coli complexed with GS-menadione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, L-gamma-glutamyl-S-(3-methyl-1,4-dioxo-1,4-dihydronaphthalen-2-yl)-L-cysteinylglycine, SULFATE ION, ... | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.557 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
6UFA
 
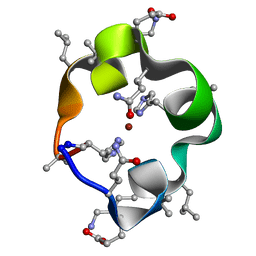 | | S4 symmetric peptide design number 1, Tim zinc-bound form | | Descriptor: | S4-1, Tim, Zinc-bound form, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-24 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UG2
 
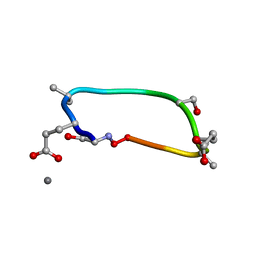 | | C2 symmetric peptide design number 1, Zappy, crystal form 2 | | Descriptor: | C2-1, Zappy, crystal form 2, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UF4
 
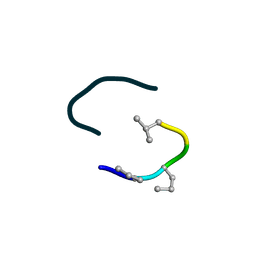 | | S2 symmetric peptide design number 4 crystal form 2, Pugsley | | Descriptor: | S2-4, Pusgley crystal form 2 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UF9
 
 | | S4 symmetric peptide design number 1, Tim apo form | | Descriptor: | S4-1, Tim apo-form, SULFATE ION | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-24 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UG6
 
 | | C3 symmetric peptide design number 1, Sporty, crystal form 2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, C3-1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDW
 
 | | S2 symmetric peptide design number 3 crystal form 2, Lurch | | Descriptor: | S2-3, Lurch crystal form 2 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UGB
 
 | | C3 symmetric peptide design number 2, Baby Basil | | Descriptor: | C3 symmetric peptide design number 2, Baby Basil, CHLORIDE ION, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
1RH1
 
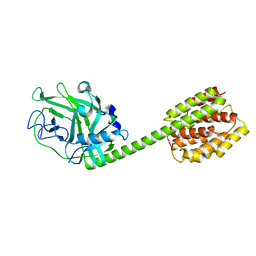 | | crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution | | Descriptor: | Colicin B | | Authors: | Hilsenbeck, J.L, Park, H, Chen, G, Youn, B, Postle, K, Kang, C. | | Deposit date: | 2003-11-13 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution
Mol.Microbiol., 51, 2004
|
|
4G5E
 
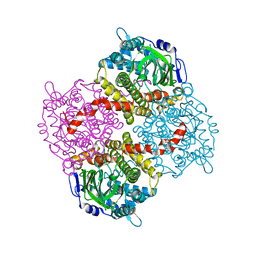 | | 2,4,6-Trichlorophenol 4-monooxygenase | | Descriptor: | 2,4,6-Trichlorophenol 4-monooxygenase | | Authors: | Hayes, R.P, Webb, B.N, Subramanian, A.K, Nissen, M, Popchock, A, Xun, L, Kang, C. | | Deposit date: | 2012-07-17 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Catalytic Differences between Two FADH(2)-Dependent Monooxygenases: 2,4,5-TCP 4-Monooxygenase (TftD) from Burkholderia cepacia AC1100 and 2,4,6-TCP 4-Monooxygenase (TcpA) from Cupriavidus necator JMP134.
Int J Mol Sci, 13, 2012
|
|
4G0I
 
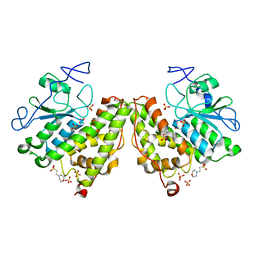 | | Glutathionyl-Hydroquinone Reductase, YqjG of Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, protein yqjG | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
1RF6
 
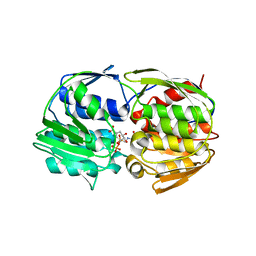 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase in S3P-GLP Bound State | | Descriptor: | 5-enolpyruvylshikimate-3-phosphate synthase, GLYPHOSATE, SHIKIMATE-3-PHOSPHATE | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
6UD9
 
 | | S2 symmetric peptide design number 2, Morticia | | Descriptor: | S2-2, Morticia | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UCX
 
 | | S2 symmetric peptide design number 1, Wednesday | | Descriptor: | S2-1, Wednesday, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDZ
 
 | | S2 symmetric peptide design number 4 crystal form 1, Pugsley | | Descriptor: | S2-4, Pusgley crystal form 1, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
4RNS
 
 | | PcpR inducer binding domain (apo-form) | | Descriptor: | PCP degradation transcriptional activation protein | | Authors: | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | Deposit date: | 2014-10-25 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
4RPO
 
 | | PcpR inducer binding domain (Complex with 2,4,6-trichlorophenol) | | Descriptor: | 2,4,6-trichlorophenol, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
4RPN
 
 | | PcpR inducer binding domain complex with pentachlorophenol | | Descriptor: | PCP degradation transcriptional activation protein, PENTACHLOROPHENOL | | Authors: | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
1RF5
 
 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase in Unliganded State | | Descriptor: | 5-enolpyruvylshikimate-3-phosphate synthase | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
4FQU
 
 | |
6UF7
 
 | | S2 symmetric peptide design number 5, Uncle Fester | | Descriptor: | S2-5, Uncle Fester | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
