4KE4
 
 | | Elucidation of the structure and reaction mechanism of Sorghum bicolor hydroxycinnamoyltransferase and its structural relationship to other CoA-dependent transferases and synthases | | Descriptor: | GLYCEROL, Hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase, TETRAETHYLENE GLYCOL | | Authors: | Walker, A.M, Hayes, R.P, Youn, B, Vermerris, W, Sattler, S.E, Kang, C. | | Deposit date: | 2013-04-25 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme a-dependent transferases and synthases.
Plant Physiol., 162, 2013
|
|
4LBP
 
 | | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG) from Burkholderia phenoliruptrix AC1100: Complex with 2,5-dihydroxybenzoquinone | | Descriptor: | 2,5-dihydroxycyclohexa-2,5-diene-1,4-dione, 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG) | | Authors: | Hayes, R.P, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-28 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Catalytic Mechanism of 5-Chlorohydroxyhydroquinone Dehydrochlorinase from the YCII Superfamily of Largely Unknown Function.
J.Biol.Chem., 288, 2013
|
|
1IMS
 
 | | MOLECULAR STRUCTURE OF THE HALOGENATED ANTI-CANCER DRUG IODODOXORUBICIN COMPLEXED WITH D(TGTACA) AND D(CGATCG) | | Descriptor: | 4'-DEOXY-4'-IODODOXORUBICIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Berger, I, Su, L, Spitzner, J.R, Kang, C, Burke, T.G, Rich, A. | | Deposit date: | 1995-10-23 | | Release date: | 1996-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular structure of the halogenated anti-cancer drug iododoxorubicin complexed with d(TGTACA) and d(CGATCG).
Nucleic Acids Res., 23, 1995
|
|
2BGM
 
 | | X-Ray structure of ternary-Secoisolariciresinol Dehydrogenase | | Descriptor: | MATAIRESINOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE | | Authors: | Youn, B, Moinuddin, S.G, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Apo-Form and Binary/Ternary Complexes of Podophyllum Secoisolariciresinol Dehydrogenase, an Enzyme Involved in Formation of Health-Protecting and Plant Defense Lignans
J.Biol.Chem., 280, 2005
|
|
1IMR
 
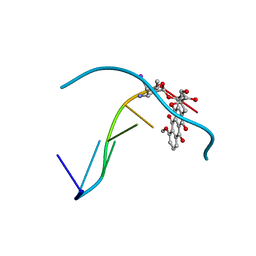 | | MOLECULAR STRUCTURE OF THE HALOGENATED ANTI-CANCER DRUG IODODOXORUBICIN COMPLEXED WITH D(TGTACA) AND D(CGATCG) | | Descriptor: | 4'-DEOXY-4'-IODODOXORUBICIN, DNA (5'-D(*TP*GP*TP*AP*CP*A)-3') | | Authors: | Berger, I, Su, L, Spitzner, J.R, Kang, C, Burke, T.G, Rich, A. | | Deposit date: | 1995-10-23 | | Release date: | 1996-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular structure of the halogenated anti-cancer drug iododoxorubicin complexed with d(TGTACA) and d(CGATCG).
Nucleic Acids Res., 23, 1995
|
|
1RF6
 
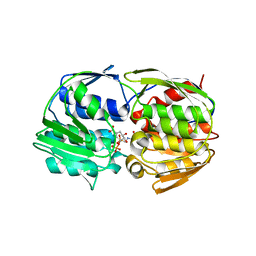 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase in S3P-GLP Bound State | | Descriptor: | 5-enolpyruvylshikimate-3-phosphate synthase, GLYPHOSATE, SHIKIMATE-3-PHOSPHATE | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
2BGL
 
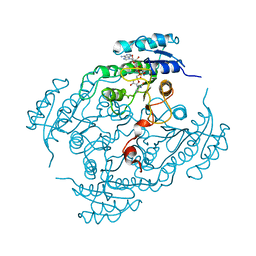 | | X-Ray structure of binary-Secoisolariciresinol Dehydrogenase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE | | Authors: | Youn, B, Moinuddin, S.G, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Apo-Form and Binary/Ternary Complexes of Podophyllum Secoisolariciresinol Dehydrogenase, an Enzyme Involved in Formation of Health-Protecting and Plant Defense Lignans
J.Biol.Chem., 280, 2005
|
|
3ULL
 
 | |
1S66
 
 | | Crystal structure of heme domain of direct oxygen sensor from E. coli | | Descriptor: | Hypothetical protein yddU, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, H.J, Suquet, C, Satterlee, J.D, Kang, C.H. | | Deposit date: | 2004-01-22 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into signal transduction involving PAS domain oxygen-sensing heme proteins from the X-ray crystal structure of Escherichia coli Dos heme domain (Ec DosH)
Biochemistry, 43, 2004
|
|
1S3O
 
 | |
1S67
 
 | | Crystal structure of heme domain of direct oxygen sensor from E. coli | | Descriptor: | Hypothetical protein yddU, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, H.J, Suquet, C, Satterlee, J.D, Kang, C.H. | | Deposit date: | 2004-01-22 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into signal transduction involving PAS domain oxygen-sensing heme proteins from the X-ray crystal structure of Escherichia coli Dos heme domain (Ec DosH)
Biochemistry, 43, 2004
|
|
1RF4
 
 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase, Tetrahedral intermediate Bound State | | Descriptor: | (3R,4S,5R)-5-{[(1R)-1-CARBOXY-2-FLUORO-1-(PHOSPHONOOXY)ETHYL]OXY}-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 5-enolpyruvylshikimate-3-phosphate synthase | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
4LBI
 
 | |
2NDJ
 
 | | Structural Basis for KCNE3 and Estrogen Modulation of the KCNQ1 Channel | | Descriptor: | Potassium voltage-gated channel subfamily E member 3 | | Authors: | Sanders, C.R, Van Horn, W.D, Kroncke, B.M, Sisco, N.J, Meiler, J, Vanoye, C.G, Song, Y, Nannemann, D.P, Welch, R.C, Kang, C, Smith, J, George, A.L. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for KCNE3 modulation of potassium recycling in epithelia.
Sci Adv, 2, 2016
|
|
1RF5
 
 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase in Unliganded State | | Descriptor: | 5-enolpyruvylshikimate-3-phosphate synthase | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
1T9P
 
 | | Crystal Structure of V44A, G45P Cp Rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Park, I.Y, Eidsness, M.K, Lin, I.J, Gebel, E.B, Youn, B, Harley, J.L, Machonkin, T.E, Frederick, R.O, Markley, J.L, Smith, E.T, Ichiye, T, Kang, C. | | Deposit date: | 2004-05-18 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic studies of V44 mutants of Clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential.
Proteins, 57, 2004
|
|
1T9O
 
 | | Crystal Structure of V44G Cp Rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Park, I.Y, Eidsness, M.K, Lin, I.J, Gebel, E.B, Youn, B, Harley, J.L, Machonkin, T.E, Frederick, R.O, Markley, J.L, Smith, E.T, Ichiye, T, Kang, C. | | Deposit date: | 2004-05-18 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies of V44 mutants of Clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential.
Proteins, 57, 2004
|
|
4LTD
 
 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - apo form | | Descriptor: | NADH-dependent FMN reductase, PHOSPHATE ION, SULFATE ION | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
1T9Q
 
 | | Crystal Structure of V44L Cp Rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Park, I.Y, Eidsness, M.K, Lin, I.J, Gebel, E.B, Youn, B, Harley, J.L, Machonkin, T.E, Frederick, R.O, Markley, J.L, Smith, E.T, Ichiye, T, Kang, C. | | Deposit date: | 2004-05-18 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies of V44 mutants of Clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential.
Proteins, 57, 2004
|
|
4G5E
 
 | | 2,4,6-Trichlorophenol 4-monooxygenase | | Descriptor: | 2,4,6-Trichlorophenol 4-monooxygenase | | Authors: | Hayes, R.P, Webb, B.N, Subramanian, A.K, Nissen, M, Popchock, A, Xun, L, Kang, C. | | Deposit date: | 2012-07-17 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Catalytic Differences between Two FADH(2)-Dependent Monooxygenases: 2,4,5-TCP 4-Monooxygenase (TftD) from Burkholderia cepacia AC1100 and 2,4,6-TCP 4-Monooxygenase (TcpA) from Cupriavidus necator JMP134.
Int J Mol Sci, 13, 2012
|
|
1SJI
 
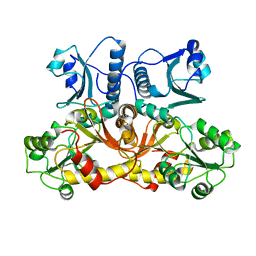 | | Comparing skeletal and cardiac calsequestrin structures and their calcium binding: a proposed mechanism for coupled calcium binding and protein polymerization | | Descriptor: | Calsequestrin, cardiac muscle isoform | | Authors: | Park, H.J, Park, I.Y, Kim, E.J, Youn, B, Fields, K, Dunker, A.K, Kang, C.H. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparing skeletal and cardiac calsequestrin structures and their calcium binding: a proposed mechanism for coupled calcium binding and protein polymerization.
J.Biol.Chem., 279, 2004
|
|
4LTM
 
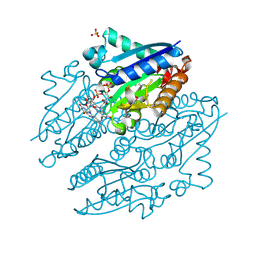 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADH-dependent FMN reductase, SULFATE ION | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
1RH1
 
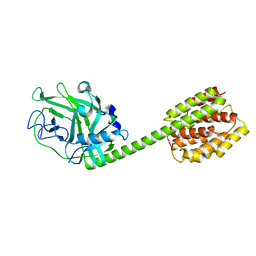 | | crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution | | Descriptor: | Colicin B | | Authors: | Hilsenbeck, J.L, Park, H, Chen, G, Youn, B, Postle, K, Kang, C. | | Deposit date: | 2003-11-13 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution
Mol.Microbiol., 51, 2004
|
|
2J3K
 
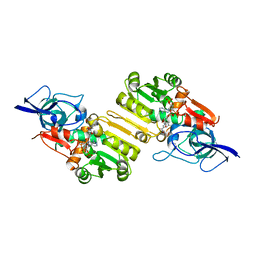 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex II | | Descriptor: | (2E,4R)-4-HYDROXYNON-2-ENAL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
2J3I
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Binary Complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT OXIDOREDUCTASE P1 | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
