3K86
 
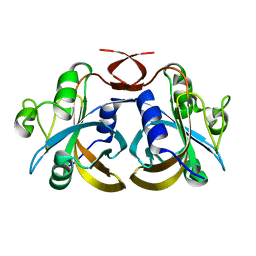 | |
6KYC
 
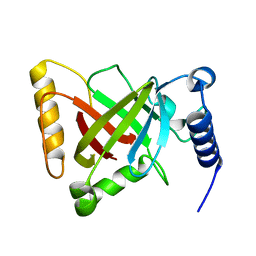 | | Structure of the S207A mutant of Clostridium difficile sortase B | | Descriptor: | Putative peptidase C60B, sortase B | | Authors: | Kang, C.Y, Huang, I.H, Wu, T.Y, Chang, J.C, Hsiao, Y.Y, Cheng, C.H, Tsai, W.J, Hsu, K.C, Wang, S.Y. | | Deposit date: | 2019-09-17 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Functional analysis ofClostridium difficilesortase B reveals key residues for catalytic activity and substrate specificity.
J.Biol.Chem., 295, 2020
|
|
6KYD
 
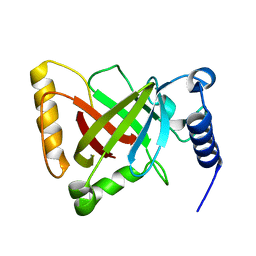 | | Structure of the R217A mutant of Clostridium difficile sortase B | | Descriptor: | Putative peptidase C60B, sortase B | | Authors: | Kang, C.Y, Huang, I.H, Wu, T.Y, Chang, J.C, Hsiao, Y.Y, Cheng, C.H, Tsai, W.J, Hsu, K.C, Wang, S.Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Functional analysis ofClostridium difficilesortase B reveals key residues for catalytic activity and substrate specificity.
J.Biol.Chem., 295, 2020
|
|
2OFN
 
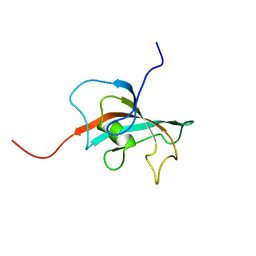 | | Solution structure of FK506-binding domain (FKBD)of FKBP35 from Plasmodium falciparum | | Descriptor: | 70 kDa peptidylprolyl isomerase, putative | | Authors: | Kang, C.B, Ye, H, Yoon, H.R, Yoon, H.S. | | Deposit date: | 2007-01-04 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FK506 binding domain (FKBD) of Plasmodium falciparum FK506 binding protein 35 (PfFKBP35).
Proteins, 70, 2007
|
|
3K88
 
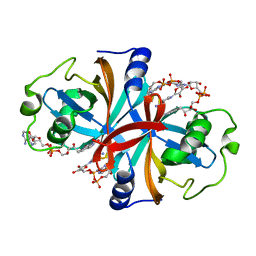 | | Crystal structure of NADH:FAD oxidoreductase (TftC) - FAD, NADH complex | | Descriptor: | Chlorophenol-4-monooxygenase component 1, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kang, C, Webb, B.N. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of chlorophenol 4-monooxygenase (TftD) and NADH:FAD oxidoreductase (TftC) of Burkholderia cepacia AC1100.
J.Biol.Chem., 285, 2010
|
|
3K87
 
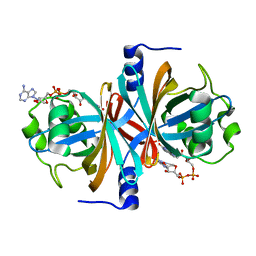 | | Crystal structure of NADH:FAD oxidoreductase (TftC) - FAD complex | | Descriptor: | Chlorophenol-4-monooxygenase component 1, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kang, C.H, Webb, B.N. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of chlorophenol 4-monooxygenase (TftD) and NADH:FAD oxidoreductase (TftC) of Burkholderia cepacia AC1100.
J.Biol.Chem., 285, 2010
|
|
1D59
 
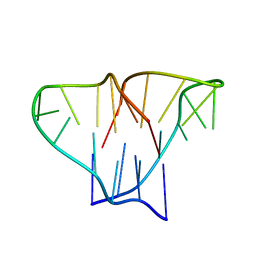 | | CRYSTAL STRUCTURE OF 4-STRANDED OXYTRICHA TELOMERIC DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3') | | Authors: | Kang, C, Zhang, X, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1992-02-25 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of four-stranded Oxytricha telomeric DNA.
Nature, 356, 1992
|
|
8QU2
 
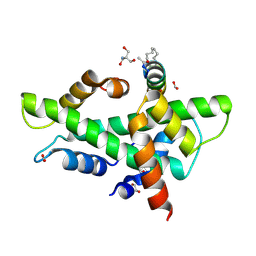 | | NF-YB/C Heterodimer in Complex with a 16-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Durukan, C, Arbore, F, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
2MF9
 
 | | Solution structure of the N-terminal domain of human FKBP38 (FKBP38NTD) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP8 | | Authors: | Kang, C, Ye, H, Simon, B, Sattler, M, Yoon, H.S. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Functional role of the flexible N-terminal extension of FKBP38 in catalysis.
Sci Rep, 3, 2013
|
|
1REG
 
 | |
191D
 
 | | CRYSTAL STRUCTURE OF INTERCALATED FOUR-STRANDED D(C3T) | | Descriptor: | DNA (5'-D(*CP*CP*CP*T)-3'), SODIUM ION | | Authors: | Kang, C, Berger, I, Lockshin, C, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1994-09-29 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of intercalated four-stranded d(C3T) at 1.4 angstroms resolution.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
200D
 
 | | STABLE LOOP IN THE CRYSTAL STRUCTURE OF THE INTERCALATED FOUR-STRANDED CYTOSINE-RICH METAZOAN TELOMERE | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*CP*C)-3') | | Authors: | Kang, C, Berger, I, Lockshin, C, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1995-02-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Stable loop in the crystal structure of the intercalated four-stranded cytosine-rich metazoan telomere.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
2K21
 
 | | NMR structure of human KCNE1 in LMPG micelles at pH 6.0 and 40 degree C | | Descriptor: | Potassium voltage-gated channel subfamily E member | | Authors: | Kang, C, Tian, C, Sonnichsen, F.D, Smith, J.A, Meiler, J, George, A.L, Vanoye, C.G, Sanders, C.R, Kim, H. | | Deposit date: | 2008-03-19 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel.
Biochemistry, 47, 2008
|
|
1VR2
 
 | | HUMAN VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 2 (KDR) KINASE DOMAIN | | Descriptor: | PROTEIN (VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR KINASE) | | Authors: | Mctigue, M, Wickersham, J, Pinko, C, Showalter, R, Parast, C, Tempczyk-Russell, A, Gehring, M, Mroczkowski, B, Kan, C, Villafranca, J, Appelt, K. | | Deposit date: | 1998-12-03 | | Release date: | 2000-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the kinase domain of human vascular endothelial growth factor receptor 2: a key enzyme in angiogenesis.
Structure Fold.Des., 7, 1999
|
|
1IA8
 
 | | THE 1.7 A CRYSTAL STRUCTURE OF HUMAN CELL CYCLE CHECKPOINT KINASE CHK1 | | Descriptor: | CHK1 CHECKPOINT KINASE, SULFATE ION | | Authors: | Chen, P, Luo, C, Deng, Y, Ryan, K, Register, J, Margosiak, S, Tempczyk-Russell, A, Nguyen, B, Myers, P, Lundgren, K, Chen Kan, C.-C, O'Connor, P.M. | | Deposit date: | 2001-03-22 | | Release date: | 2001-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation.
Cell(Cambridge,Mass.), 100, 2000
|
|
1CDE
 
 | | STRUCTURES OF APO AND COMPLEXED ESCHERICHIA COLI GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | 5-DEAZAFOLIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, PHOSPHORIBOSYL-GLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Almassy, R.J, Janson, C.A, Kan, C.-C, Hostomska, Z. | | Deposit date: | 1992-05-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of apo and complexed Escherichia coli glycinamide ribonucleotide transformylase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1CDD
 
 | | STRUCTURES OF APO AND COMPLEXED ESCHERICHIA COLI GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | PHOSPHATE ION, PHOSPHORIBOSYL-GLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Almassy, R.J, Janson, C.A, Kan, C.-C, Hostomska, Z. | | Deposit date: | 1992-05-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of apo and complexed Escherichia coli glycinamide ribonucleotide transformylase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
8DJR
 
 | | Cytosolic ascorbate peroxidase from Sorghum bicolor | | Descriptor: | L-ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A sorghum ascorbate peroxidase with four binding sites has activity against ascorbate and phenylpropanoids.
Plant Physiol., 192, 2023
|
|
8DJX
 
 | | Cytosolic ascorbate peroxidase from Sorghum bicolor - Compound II | | Descriptor: | L-ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A sorghum ascorbate peroxidase with four binding sites has activity against ascorbate and phenylpropanoids.
Plant Physiol., 192, 2023
|
|
8DJU
 
 | | Cytosolic ascorbate peroxidase from Sorghum bicolor - bicyclic dehydroascorbic acid complex | | Descriptor: | (3aS,6S,6aR)-3,3,3a,6-tetrahydroxytetrahydrofuro[3,2-b]furan-2(3H)-one (non-preferred name), L-ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A sorghum ascorbate peroxidase with four binding sites has activity against ascorbate and phenylpropanoids.
Plant Physiol., 192, 2023
|
|
8DJW
 
 | |
9B1R
 
 | | Functional implication of the homotrimeric multidomain vacuolar sorting receptor 1 from Arabidopsis thaliana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Vacuolar-sorting receptor 1 | | Authors: | Park, H, Youn, B, Park, D.J, Puthanveettil, S.V, Kang, C. | | Deposit date: | 2024-03-13 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Functional implication of the homotrimeric multidomain vacuolar sorting receptor 1 (VSR1) from Arabidopsis thaliana.
Sci Rep, 14, 2024
|
|
8FEM
 
 | | Panicum vigratum Dihydroflavonol 4-reductase complexed with NADP | | Descriptor: | Dihydroflavonol 4-Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FET
 
 | | Flavanone 4-Reductase from Sorghum bicolor-NADP(H) complex | | Descriptor: | 3-deoxyanthocyanidin synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEV
 
 | | Flavanone 4-Reductase from Sorghum bicolor-NADP(H) and dihydroquercetin complex | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 3-deoxyanthocyanidin synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
