1JF4
 
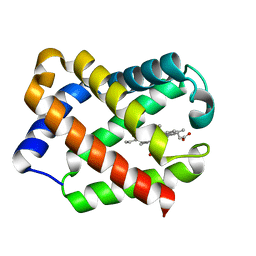 | | Crystal Structure Of Component IV Glycera Dibranchiata Monomeric Hemoglobin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, monomer hemoglobin component IV | | Authors: | Park, H.J, Yang, C, Treff, N, Satterlee, J.D, Kang, C.H. | | Deposit date: | 2001-06-20 | | Release date: | 2002-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of Unligated and CN-Ligated Glycera dibranchiata Monomer
Ferric Hemoglobin Components III and IV
Proteins, 49, 2002
|
|
1JL7
 
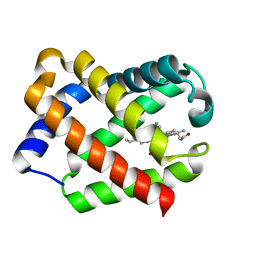 | | Crystal Structure Of CN-Ligated Component III Glycera Dibranchiata Monomeric Hemoglobin | | Descriptor: | CYANIDE ION, Monomer hemoglobin component III, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, H.J, Yang, C, Treff, N, Satterlee, J.D, Kang, C. | | Deposit date: | 2001-07-16 | | Release date: | 2002-07-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of Unligated and CN-Ligated Glycera dibranchiata Monomer Ferric Hemoglobin Components III and IV
Proteins, 49, 2002
|
|
1JF3
 
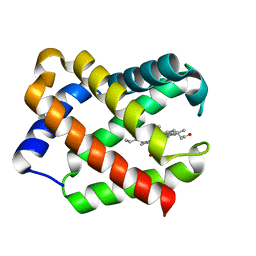 | | Crystal Structure Of Component III Glycera Dibranchiata Monomeric Hemoglobin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, monomer hemoglobin component III | | Authors: | Park, H.J, Yang, C, Treff, N, Satterlee, J.D, Kang, C.H. | | Deposit date: | 2001-06-20 | | Release date: | 2002-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of Unligated and CN-Ligated Glycera dibranchiata Monomer
Ferric Hemoglobin Components III and IV
Proteins, 49, 2002
|
|
1JL6
 
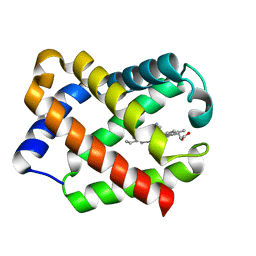 | | Crystal Structure of CN-Ligated Component IV Glycera Dibranchiata Monomeric Hemoglobin | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, monomer hemoglobin component IV | | Authors: | Park, H.J, Yang, C, Treff, N, Satterlee, J.D, Kang, C. | | Deposit date: | 2001-07-16 | | Release date: | 2002-07-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of Unligated and CN-Ligated Glycera dibranchiata Monomer Ferric Hemoglobin Components III and IV
Proteins, 49, 2002
|
|
1T9O
 
 | | Crystal Structure of V44G Cp Rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Park, I.Y, Eidsness, M.K, Lin, I.J, Gebel, E.B, Youn, B, Harley, J.L, Machonkin, T.E, Frederick, R.O, Markley, J.L, Smith, E.T, Ichiye, T, Kang, C. | | Deposit date: | 2004-05-18 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies of V44 mutants of Clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential.
Proteins, 57, 2004
|
|
2ONG
 
 | | Crystal Structure of of limonene synthase with 2-fluorogeranyl diphosphate (FGPP). | | Descriptor: | (1S)-1-[(1R)-1-FLUOROETHYL]-1,5-DIMETHYLHEXYL TRIHYDROGEN DIPHOSPHATE, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4S-limonene synthase, ... | | Authors: | Hyatt, D.C, Youn, B, Croteau, R, Kang, C. | | Deposit date: | 2007-01-23 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of limonene synthase, a simple model for terpenoid cyclase catalysis.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2LAO
 
 | | THREE-DIMENSIONAL STRUCTURES OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN WITH AND WITHOUT A LIGAND | | Descriptor: | LYSINE, ARGININE, ORNITHINE-BINDING PROTEIN | | Authors: | Kim, S.-H, Oh, B.-H, Kang, C.-H. | | Deposit date: | 1993-02-25 | | Release date: | 1994-06-22 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without a ligand.
J.Biol.Chem., 268, 1993
|
|
2ONH
 
 | | Crystal Structure of of limonene synthase with 2-fluorolinalyl diphosphate(FLPP) | | Descriptor: | (1S)-1-[(1S)-1-FLUOROETHYL]-1,5-DIMETHYLHEXYL TRIHYDROGEN DIPHOSPHATE, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4S-limonene synthase, ... | | Authors: | Hyatt, D.C, Youn, B, Croteau, R, Kang, C. | | Deposit date: | 2007-01-24 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of limonene synthase, a simple model for terpenoid cyclase catalysis.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1T9Q
 
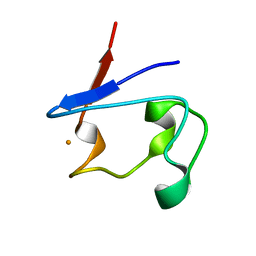 | | Crystal Structure of V44L Cp Rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Park, I.Y, Eidsness, M.K, Lin, I.J, Gebel, E.B, Youn, B, Harley, J.L, Machonkin, T.E, Frederick, R.O, Markley, J.L, Smith, E.T, Ichiye, T, Kang, C. | | Deposit date: | 2004-05-18 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies of V44 mutants of Clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential.
Proteins, 57, 2004
|
|
2J3H
 
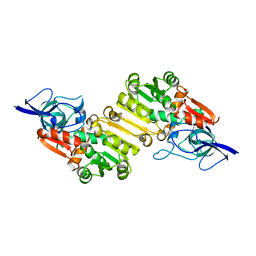 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Apo form | | Descriptor: | NADP-DEPENDENT OXIDOREDUCTASE P1 | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
3V1W
 
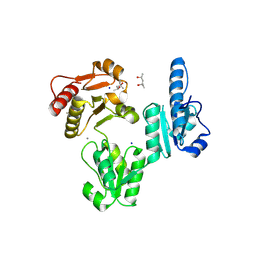 | | Molecular Basis for Multiple Ligand Binding of Calsequestrin and Potential Inhibition by Caffeine and Gallocatecin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Subramanian, A.K, Nissen, M.N, Lewis, K.M, Sanchez, E.J, Muralidharan, A.K, Kang, C. | | Deposit date: | 2011-12-10 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Molecular Basis for Multiple Ligand Binding of Calsequestrin and Potential Inhibition by Caffeine and Gallocatecin
To be Published
|
|
3US3
 
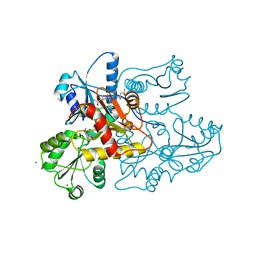 | | Recombinant rabbit skeletal calsequestrin-MPD complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Nissen, M.S, Munske, G.R, Kang, C. | | Deposit date: | 2011-11-22 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Glycosylation of skeletal calsequestrin: implications for its function.
J.Biol.Chem., 287, 2012
|
|
2J3K
 
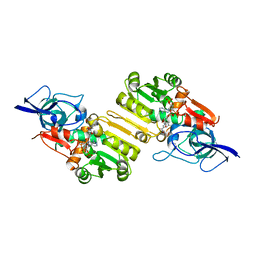 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex II | | Descriptor: | (2E,4R)-4-HYDROXYNON-2-ENAL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
2J3J
 
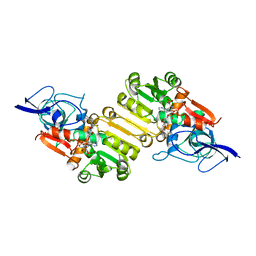 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex I | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
1RF4
 
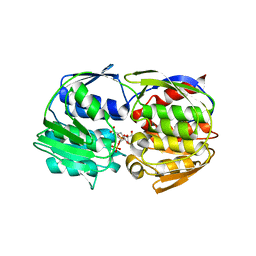 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase, Tetrahedral intermediate Bound State | | Descriptor: | (3R,4S,5R)-5-{[(1R)-1-CARBOXY-2-FLUORO-1-(PHOSPHONOOXY)ETHYL]OXY}-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 5-enolpyruvylshikimate-3-phosphate synthase | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
2J3I
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Binary Complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT OXIDOREDUCTASE P1 | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
3TRQ
 
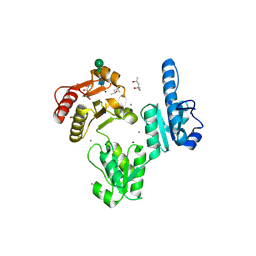 | | Crystal structure of native rabbit skeletal calsequestrin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Munske, G.R, Nissen, M.S, Kang, C. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Glycosylation of Skeletal Calsequestrin: IMPLICATIONS FOR ITS FUNCTION.
J.Biol.Chem., 287, 2012
|
|
3TRP
 
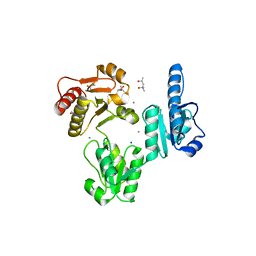 | | Crystal structure of recombinant rabbit skeletal calsequestrin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Munske, G.R, Nissen, M.S, Kang, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8817 Å) | | Cite: | High-capacity Ca2+ Binding of Human Skeletal Calsequestrin.
J.Biol.Chem., 287, 2012
|
|
1QYC
 
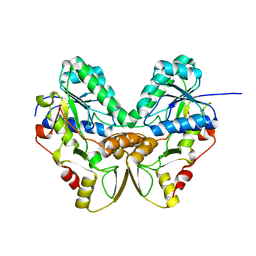 | | Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases, and their relationship to isoflavone reductases | | Descriptor: | phenylcoumaran benzylic ether reductase PT1 | | Authors: | Min, T, Kasahara, H, Bedgar, D.L, Youn, B, Lawrence, P.K, Gang, D.R, Halls, S.C, Park, H, Hilsenbeck, J.L, Davin, L.B, Kang, C. | | Deposit date: | 2003-09-10 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases and their relationship to isoflavone reductases.
J.Biol.Chem., 278, 2003
|
|
1S67
 
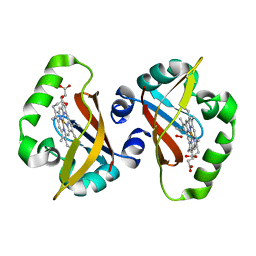 | | Crystal structure of heme domain of direct oxygen sensor from E. coli | | Descriptor: | Hypothetical protein yddU, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, H.J, Suquet, C, Satterlee, J.D, Kang, C.H. | | Deposit date: | 2004-01-22 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into signal transduction involving PAS domain oxygen-sensing heme proteins from the X-ray crystal structure of Escherichia coli Dos heme domain (Ec DosH)
Biochemistry, 43, 2004
|
|
2CF5
 
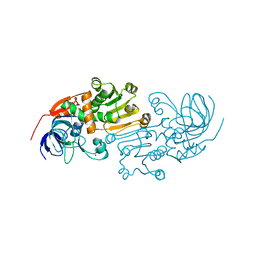 | | Crystal Structures of the Arabidopsis Cinnamyl Alcohol Dehydrogenases, AtCAD5 | | Descriptor: | CINNAMYL ALCOHOL DEHYDROGENASE, ZINC ION | | Authors: | Youn, B, Camacho, R, Moinuddin, S, Lee, C, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-02-16 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures and Catalytic Mechanisms of the Arabidopsis Cinnamyl Alcohol Dehydrogenases Atcad5 and Atcad4
Org.Biomol.Chem., 4, 2006
|
|
2CF6
 
 | | Crystal Structures of the Arabidopsis Cinnamyl Alcohol Dehydrogenases AtCAD5 | | Descriptor: | CINNAMYL ALCOHOL DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Youn, B, Camacho, R, Moinuddin, S.G, Lee, C, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-02-16 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures and Catalytic Mechanisms of the Arabidopsis Cinnamyl Alcohol Dehydrogenases Atcad5 and Atcad4
Org.Biomol.Chem., 4, 2006
|
|
1S3O
 
 | |
1RF5
 
 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase in Unliganded State | | Descriptor: | 5-enolpyruvylshikimate-3-phosphate synthase | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
1RF6
 
 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase in S3P-GLP Bound State | | Descriptor: | 5-enolpyruvylshikimate-3-phosphate synthase, GLYPHOSATE, SHIKIMATE-3-PHOSPHATE | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
