5IEL
 
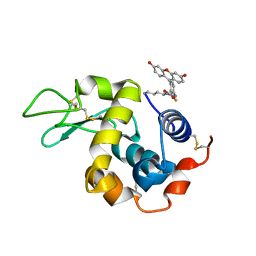 | | Structure of Lysozyme labeled with fluorescein isothiocyanate (FITC) at 1.15 Angstroms resolution | | Descriptor: | 2-(6-hydroxy-3-oxo-3H-xanthen-9-yl)-5-[(E)-(sulfanylmethylidene)amino]benzoic acid, Lysozyme C | | Authors: | Kachalova, G.S, Vlaskina, A.V, Popov, A.P, Simanovskaia, A.A, Krukova, M.V, Lipkin, A.V. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of Lysozyme labeled with fluorescein isothiocyanate (FITC) at 1.15 Angstroms resolution
To Be Published
|
|
2EWF
 
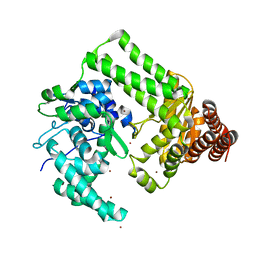 | | Crystal structure of the site-specific DNA nickase N.BspD6I | | Descriptor: | BROMIDE ION, Nicking endonuclease N.BspD6I | | Authors: | Kachalova, G.S, Bartunik, H.D, Artyukh, R.I, Rogulin, E.A, Perevyazova, T.A, Zheleznaya, L.A, Matvienko, N.I. | | Deposit date: | 2005-11-03 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of the heterodimeric type IIS restriction endonuclease R.BspD6I acting as a complex between a monomeric site-specific nickase and a catalytic subunit.
J.Mol.Biol., 384, 2008
|
|
5LIQ
 
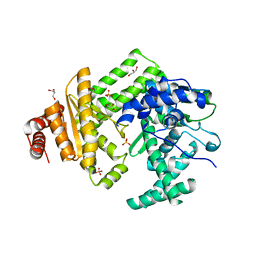 | | The structure of C160S,C508S,C578S mutant of Nt.BspD6I nicking endonuclease at 0.185 nm resolution . | | Descriptor: | GLYCEROL, Nicking endonuclease N.BspD6I, PHOSPHATE ION | | Authors: | Kachalova, G.S, Artyukh, R.I, Perevyazova, T.A, Yunusova, A.K, Popov, A.N, Bartunik, H.D, Zheleznaya, L.A. | | Deposit date: | 2016-07-15 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural features of Cysteine residues mutation of the nicking endonuclease Nt.BspD6I.
To Be Published
|
|
5LHC
 
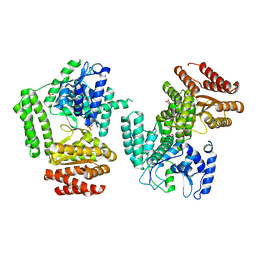 | | The structure of D456A mutant of Nt.BspD6I nicking endonuclease at 0.24 nm resolution . | | Descriptor: | GLYCEROL, Nicking endonuclease N.BspD6I, PHOSPHATE ION | | Authors: | Kachalova, G.S, Yunusova, A.K, Popov, A.N, Artyukh, R.I, Perevyazova, T.A, Bartunik, H.D, Zheleznaya, L.A. | | Deposit date: | 2016-07-10 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural implication of activity loss by D456A mutant of the nicking endonuclease Nt.BspD6I.
To Be Published
|
|
5LIZ
 
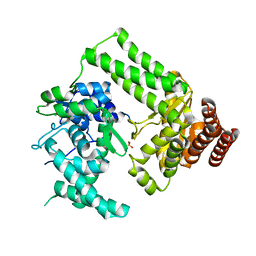 | | The structure of Nt.BspD6I nicking endonuclease with all cysteines mutated by serine residues at 0.19 nm resolution . | | Descriptor: | Nicking endonuclease N.BspD6I, PHOSPHATE ION | | Authors: | Kachalova, G.S, Artyukh, R.I, Perevyazova, T.A, Yunusova, A.K, Popov, A.N, Bartunik, H.D, Zheleznaya, L.A. | | Deposit date: | 2016-07-16 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural features of Cysteine residues mutation of the nicking endonuclease Nt.BspD6I.
To Be Published
|
|
5N9O
 
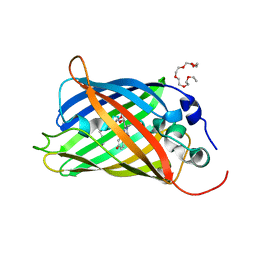 | | EGFP(enhanced green fluorescent protein) mutant - L232H | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, Green fluorescent protein | | Authors: | Kachalova, G.S, Popov, A.P, Simanovskaya, A.A, Krukova, M.V, Lipkin, A.V. | | Deposit date: | 2017-02-26 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure of EGFP(enhanced green fluorescent protein) mutant - L232H at 0.153 nm
To Be Published
|
|
3CFF
 
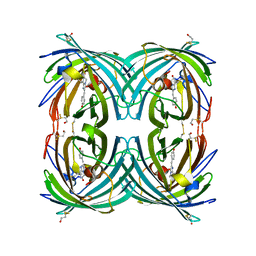 | |
3PNV
 
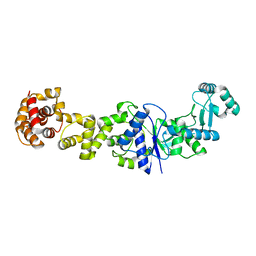 | |
3CFA
 
 | |
3CFH
 
 | |
3PNY
 
 | |
1BZP
 
 | |
1BZR
 
 | | ATOMIC RESOLUTION CRYSTAL STRUCTURE ANALYSIS OF NATIVE DEOXY AND CO MYOGLOBIN FROM SPERM WHALE AT ROOM TEMPERATURE | | Descriptor: | CARBON MONOXIDE, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kachalova, G.S, Popov, A.N, Bartunik, H.D. | | Deposit date: | 1998-11-03 | | Release date: | 1999-05-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A steric mechanism for inhibition of CO binding to heme proteins.
Science, 284, 1999
|
|
3NG7
 
 | |
4DP8
 
 | | The 1.07 Angstrom crystal structure of reduced (CuI) poplar plastocyanin A at pH 4.0 | | Descriptor: | COPPER (I) ION, Plastocyanin A, chloroplastic, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4DP5
 
 | | The 1.88 Angstrom crystal structure of oxidized (CuII) poplar plastocyanin B at pH 8.0 | | Descriptor: | COPPER (II) ION, GLYCEROL, Plastocyanin B, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4DP0
 
 | | The 1.5 Angstrom crystal structure of oxidized (CuII) poplar plastocyanin B at pH 4.0 | | Descriptor: | COPPER (II) ION, GLYCEROL, Plastocyanin B, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4DPA
 
 | | The 1.05 Angstrom crystal structure of reduced (CuI) poplar plastocyanin A at pH 6.0 | | Descriptor: | COPPER (I) ION, Plastocyanin A, chloroplastic | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
3NN0
 
 | | Complex of 6-hydroxy-L-nicotine oxidase with nicotinamide | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-23 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NK2
 
 | | Complex of 6-hydroxy-L-nicotine oxidase with dopamine | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-18 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NGC
 
 | | Complex of 6-hydroxy-L-nicotine oxidase with intermediate methylmyosmine product formed during catalytic turnover | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 5-(1-methyl-4,5-dihydro-1H-pyrrol-2-yl)pyridin-2-ol, 5-[(2S)-1-methylpyrrolidin-2-yl]pyridin-2-ol, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-11 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NH3
 
 | |
3NHO
 
 | | Complex of 6-hydroxy-L-nicotine oxidase with product bound at active site | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 1-(6-hydroxypyridin-3-yl)-4-(methylamino)butan-1-one, 6-hydroxy-L-nicotine oxidase, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NK1
 
 | | Complex of 6-hydroxy-L-nicotine oxidase with serotonin | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-18 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NN6
 
 | | Crystal structure of inhibitor-bound in active centre 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, (2R,3S,4S)-5-({[(acetylcarbamoyl)amino]methyl}[(3S,4R)-6-amino-3,4-dimethylhexyl]amino)-2,3,4-trihydroxypentyl [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), 5-[(2R)-1-methylpyrrolidin-2-yl]pyridin-2-ol, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-23 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
