4Y1F
 
 | | SAV1875-E17D | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-07 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
4Y1R
 
 | | SAV1875-cysteinesulfonic acid | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-08 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
4Y1G
 
 | | SAV1875-E17N | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-07 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
7CPA
 
 | |
1A7K
 
 | | GLYCOSOMAL GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE IN A MONOCLINIC CRYSTAL FORM | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Kim, H, Hol, W.G.J. | | Deposit date: | 1998-03-16 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Leishmania mexicana glycosomal glyceraldehyde-3-phosphate dehydrogenase in a new crystal form confirms the putative physiological active site structure.
J.Mol.Biol., 278, 1998
|
|
4MN7
 
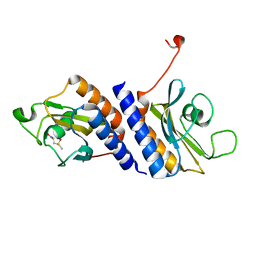 | | Structural and biochemical analysis of type II free methionine-R-sulfoxide reductase from Thermoplasma acidophilum | | Descriptor: | METHIONINE SULFOXIDE, Putative uncharacterized protein Ta0848 | | Authors: | Kim, H.S, Kwak, G.H, Lee, K.T, Jo, C.H, Hwang, K.Y, Kim, H.Y. | | Deposit date: | 2013-09-10 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical analysis of a type II free methionine-R-sulfoxide reductase from Thermoplasma acidophilum
Arch.Biochem.Biophys., 560, 2014
|
|
7EWC
 
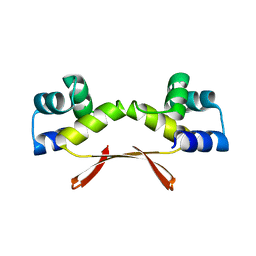 | | Mycobacterium tuberculosis HigA2 (Form I) | | Descriptor: | Putative antitoxin HigA2 | | Authors: | Kim, H.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chasing the structural diversity of the transcription regulator Mycobacterium tuberculosis HigA2.
Iucrj, 8, 2021
|
|
1R02
 
 | |
5YCR
 
 | |
5YCV
 
 | |
7VWX
 
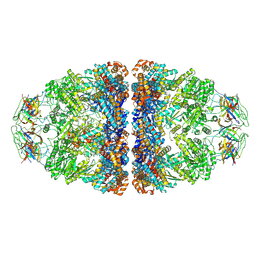 | | CryoEM structure of football-shaped GroEL:ES2 with RuBisCO | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, Ribulose bisphosphate carboxylase | | Authors: | Kim, H, Roh, S.H. | | Deposit date: | 2021-11-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Cryo-EM structures of GroEL:ES 2 with RuBisCO visualize molecular contacts of encapsulated substrates in a double-cage chaperonin.
Iscience, 25, 2022
|
|
7KWA
 
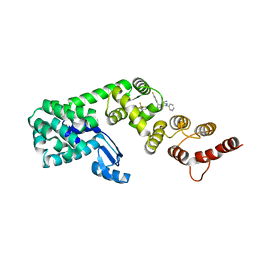 | | Structure of DCN1 bound to N-((4S,5S)-3-(aminomethyl)-7-ethyl-4-(4-fluorophenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-(trifluoromethyl)benzamide | | Descriptor: | Endolysin,DCN1-like protein 1, N-[(4S,5S)-3-(aminomethyl)-7-ethyl-4-(4-fluorophenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-(trifluoromethyl)benzamide | | Authors: | Kim, H.S, Hammill, J.T, Schulman, B.A, Guy, R.K, Scott, D.C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Improvement of Oral Bioavailability of Pyrazolo-Pyridone Inhibitors of the Interaction of DCN1/2 and UBE2M.
J.Med.Chem., 64, 2021
|
|
7EWE
 
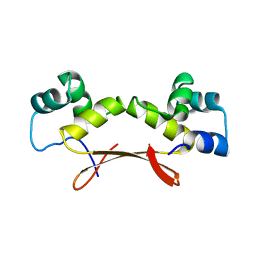 | | Mycobacterium tuberculosis HigA2 (Form III) | | Descriptor: | Putative antitoxin HigA2 | | Authors: | Kim, H.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Chasing the structural diversity of the transcription regulator Mycobacterium tuberculosis HigA2.
Iucrj, 8, 2021
|
|
7EWD
 
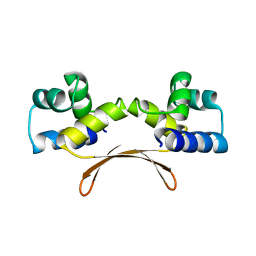 | | Mycobacterium tuberculosis HigA2 (Form II) | | Descriptor: | Putative antitoxin HigA2 | | Authors: | Kim, H.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chasing the structural diversity of the transcription regulator Mycobacterium tuberculosis HigA2.
Iucrj, 8, 2021
|
|
6KQB
 
 | | A long chain secondary alcohol dehydrogenase of Micrococcus luteus | | Descriptor: | 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Kim, H.J, Kim, J.S. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Cofactor specificity engineering of a long-chain secondary alcohol dehydrogenase from Micrococcus luteus for redox-neutral biotransformation of fatty acids.
Chem.Commun.(Camb.), 55, 2019
|
|
6KQ9
 
 | | A long chain secondary alcohol dehydrogenase of Micrococcus luteus | | Descriptor: | 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Kim, H.J, Kim, J.S. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Cofactor specificity engineering of a long-chain secondary alcohol dehydrogenase from Micrococcus luteus for redox-neutral biotransformation of fatty acids.
Chem.Commun.(Camb.), 55, 2019
|
|
1ZI8
 
 | | Crystal Structure Analysis of the dienelactone hydrolase mutant(E36D, C123S, A134S, S208G, A229V, K234R)- 1.4 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3QHM
 
 | | Crystal analysis of the complex structure, E342A-cellotetraose, of endocellulase from pyrococcus horikoshii | | Descriptor: | 458aa long hypothetical endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kim, H.-W, Ishikawa, K. | | Deposit date: | 2011-01-26 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Functional analysis of hyperthermophilic endocellulase from Pyrococcus horikoshii by crystallographic snapshots
Biochem.J., 437, 2011
|
|
5B4P
 
 | |
3QHO
 
 | | Crystal analysis of the complex structure, Y299F-cellotetraose, of endocellulase from pyrococcus horikoshii | | Descriptor: | 458aa long hypothetical endo-1,4-beta-glucanase, PHOSPHATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kim, H.-W, Ishikawa, K. | | Deposit date: | 2011-01-26 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional analysis of hyperthermophilic endocellulase from Pyrococcus horikoshii by crystallographic snapshots
Biochem.J., 437, 2011
|
|
3QHN
 
 | | Crystal analysis of the complex structure, E201A-cellotetraose, of endocellulase from pyrococcus horikoshii | | Descriptor: | 458aa long hypothetical endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kim, H.-W, Ishikawa, K. | | Deposit date: | 2011-01-26 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional analysis of hyperthermophilic endocellulase from Pyrococcus horikoshii by crystallographic snapshots
Biochem.J., 437, 2011
|
|
5GNA
 
 | |
1ZJ5
 
 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (E36D, C123S, A134S, S208G, A229V, K234R) bound with the PMS moiety of the protease inhibitor, Phenylmethylsulfonyl fluoride (PMSF)- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZIC
 
 | | Crystal Structure Analysis of the dienelactone hydrolase (C123S, R206A) mutant- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8GHA
 
 | | Hir3 Arm/Tail, Hir2 WD40, C-terminal Hpc2 | | Descriptor: | Histone promoter control protein 2, Histone transcription regulator 3, Protein HIR2 | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 2024
|
|
