1A5E
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 18 STRUCTURES | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
1A77
 
 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MAGNESIUM ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
1A76
 
 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MANGANESE (II) ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
8I9J
 
 | | The PKR and E3L complex | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase, RNA-binding protein E3 | | Authors: | Han, C.W, Kim, H.J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.39 Å) | | Cite: | Structural study of novel vaccinia virus E3L and dsRNA-dependent protein kinase complex.
Biochem.Biophys.Res.Commun., 665, 2023
|
|
1YUM
 
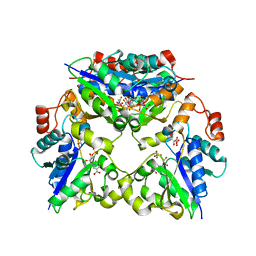 | | Crystal Structure of Nicotinic Acid Mononucleotide Adenylyltransferase from Pseudomonas aeruginosa | | Descriptor: | 'Probable nicotinate-nucleotide adenylyltransferase, CITRIC ACID, NICOTINATE MONONUCLEOTIDE | | Authors: | Yoon, H.J, Kim, H.L, Mikami, B, Suh, S.W. | | Deposit date: | 2005-02-14 | | Release date: | 2005-11-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of nicotinic acid mononucleotide adenylyltransferase from Pseudomonas aeruginosa in its Apo and substrate-complexed forms reveals a fully open conformation
J.Mol.Biol., 351, 2005
|
|
4GCV
 
 | |
3FXI
 
 | | Crystal structure of the human TLR4-human MD-2-E.coli LPS Ra complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-HYDROXY-TETRADECANOIC ACID, ... | | Authors: | Park, B.S, Song, D.H, Kim, H.M, Lee, J.-O. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex
Nature, 458, 2009
|
|
3GN4
 
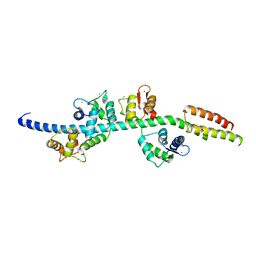 | | Myosin lever arm | | Descriptor: | CALCIUM ION, Calmodulin, MAGNESIUM ION, ... | | Authors: | Mukherjea, M, Llinas, P, Kim, H, Travaglia, M, Safer, D, Zong, A.B, Menetrey, J, Franzini-Armstrong, C, Selvin, P.R, Houdusse, A, Sweeney, H.L. | | Deposit date: | 2009-03-16 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Myosin VI dimerization triggers an unfolding of a three-helix bundle in order to extend its reach
Mol.Cell, 35, 2009
|
|
1PVE
 
 | |
7WBM
 
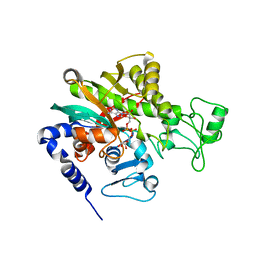 | | Crystal structure of Legionella pneumophila effector protein Lpg0081 | | Descriptor: | Lpg0081, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Lee, J, Kim, H, Oh, B.H. | | Deposit date: | 2021-12-17 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reversible modification of mitochondrial ADP/ATP translocases by paired Legionella effector proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WBK
 
 | |
2KEF
 
 | | Solution NMR structures of human hepcidin at 325K | | Descriptor: | Hepcidin | | Authors: | Jordan, J.B, Poppe, L, Hainu, M, Arvedson, T, Syed, R, Li, V, Kohno, H, Kim, H, Miranda, L.P, Cheetham, J, Sasu, B.J. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Hepcidin revisited, disulfide connectivity, dynamics, and structure.
J.Biol.Chem., 284, 2009
|
|
2DS8
 
 | | Structure of the ZBD-XB complex | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, SspB-tail peptide, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Kim, H.W, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
1YUL
 
 | | Crystal Structure of Nicotinic Acid Mononucleotide Adenylyltransferase from Pseudomonas aeruginosa | | Descriptor: | CITRIC ACID, Probable nicotinate-nucleotide adenylyltransferase | | Authors: | Yoon, H.J, Kim, H.L, Mikami, B, Suh, S.W. | | Deposit date: | 2005-02-14 | | Release date: | 2005-11-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of nicotinic acid mononucleotide adenylyltransferase from Pseudomonas aeruginosa in its Apo and substrate-complexed forms reveals a fully open conformation
J.Mol.Biol., 351, 2005
|
|
3A3X
 
 | | Structure of OpdA mutant (G60A/A80V/R118Q/K185R/Q206P/D208G/I260T/G273S) | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | Ollis, D.L, Tawfik, D.S, Schenk, G, Jackson, C.J, Foo, J.L, Tokuriki, N, Afriat, L, Carr, P.D, Kim, H.K. | | Deposit date: | 2009-06-23 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase
Proc.Natl.Acad.Sci.USA, 2009
|
|
3SJF
 
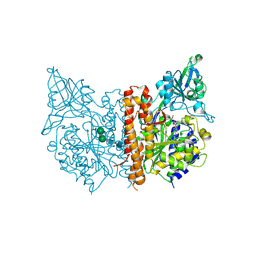 | | X-ray structure of human glutamate carboxypeptidase II in complex with a urea-based inhibitor (A25) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | Deposit date: | 2011-06-21 | | Release date: | 2011-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
3SJG
 
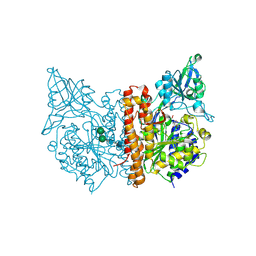 | | Human glutamate carboxypeptidase II (E424A inactive mutant ) in complex with N-acetyl-aspartyl-aminooctanoic acid | | Descriptor: | (2S)-2-[(N-acetyl-L-alpha-aspartyl)amino]nonanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | Deposit date: | 2011-06-21 | | Release date: | 2011-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
3SJE
 
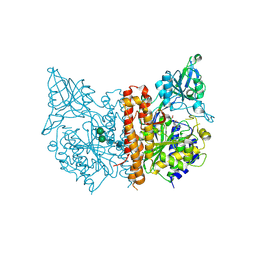 | | X-ray structure of human glutamate carboxypeptidase II (the E424A inactive mutant) in complex with N-acetyl-aspartyl-aminononanoic acid | | Descriptor: | (2S)-2-[(N-acetyl-L-alpha-aspartyl)amino]nonanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | Deposit date: | 2011-06-21 | | Release date: | 2011-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
3SJX
 
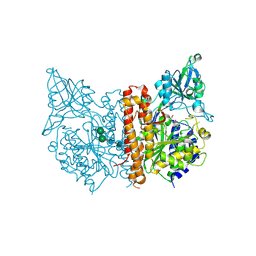 | | X-ray structure of human glutamate carboxypeptidase II (the E424A inactive mutant) in complex with N-acetyl-aspartyl-methionine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | Deposit date: | 2011-06-22 | | Release date: | 2011-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
3KYG
 
 | | Crystal structure of VCA0042 (L135R) complexed with c-di-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Putative uncharacterized protein VCA0042 | | Authors: | Ryu, K.S, Ko, J, Kim, H, Choi, B.S. | | Deposit date: | 2009-12-06 | | Release date: | 2010-04-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of PP4397 Reveals the Molecular Basis for Different c-di-GMP Binding Modes by Pilz Domain Proteins.
J.Mol.Biol., 398, 2010
|
|
2O6R
 
 | |
2O6S
 
 | |
2O6Q
 
 | |
3QJM
 
 | | Structural flexibility of Shank PDZ domain is important for its binding to different ligands | | Descriptor: | Beta-PIX, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Lee, J.H, Park, H, Park, S.J, Kim, H.J, Eom, S.H. | | Deposit date: | 2011-01-30 | | Release date: | 2011-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | The structural flexibility of the shank1 PDZ domain is important for its binding to different ligands
Biochem.Biophys.Res.Commun., 407, 2011
|
|
1Y0Q
 
 | | Crystal structure of an active group I ribozyme-product complex | | Descriptor: | 5'-R(*GP*CP*UP*U)-3', Group I ribozyme, MAGNESIUM ION, ... | | Authors: | Golden, B.L, Kim, H, Chase, E. | | Deposit date: | 2004-11-16 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of a phage Twort group I ribozyme-product complex
Nat.Struct.Mol.Biol., 12, 2005
|
|
