3MWM
 
 | |
6V5K
 
 | |
3L18
 
 | |
3LPS
 
 | | Crystal structure of parE | | Descriptor: | NOVOBIOCIN, Topoisomerase IV subunit B | | Authors: | Jung, H.Y, Heo, Y.-S. | | Deposit date: | 2010-02-05 | | Release date: | 2011-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of parE
To be Published
|
|
1DEP
 
 | | MEMBRANE PROTEIN, NMR, 1 STRUCTURE | | Descriptor: | T345-359 | | Authors: | Jung, H, Schnackerz, K.D. | | Deposit date: | 1995-08-23 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR and circular dichroism studies of synthetic peptides derived from the third intracellular loop of the beta-adrenoceptor.
FEBS Lett., 358, 1995
|
|
4M2X
 
 | |
2KSG
 
 | |
3LNU
 
 | |
3LPX
 
 | | Crystal structure of GyrA | | Descriptor: | DNA gyrase, A subunit | | Authors: | Jung, H.Y, Heo, Y.S. | | Deposit date: | 2010-02-07 | | Release date: | 2011-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of GyrA
To be Published
|
|
8KCA
 
 | |
2JMK
 
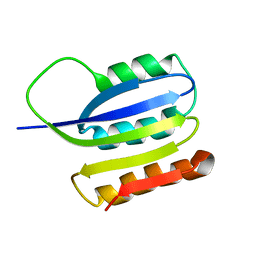 | | Solution structure of ta0956 | | Descriptor: | Hypothetical protein Ta0956 | | Authors: | Koo, B, Jung, J, Jung, H, Nam, H, Kim, Y, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2006-11-20 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical novel-fold protein TA0956 from Thermoplasma acidophilum
Proteins, 69, 2007
|
|
1ZKJ
 
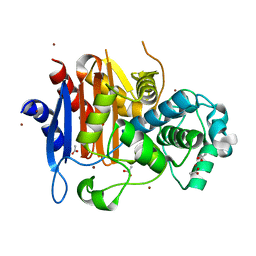 | | Structural Basis for the Extended Substrate Spectrum of CMY-10, a Plasmid-Encoded Class C beta-lactamase | | Descriptor: | ACETIC ACID, ZINC ION, extended-spectrum beta-lactamase | | Authors: | Cha, S.S, Jung, H.I, An, Y.J, Lee, S.H. | | Deposit date: | 2005-05-03 | | Release date: | 2006-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C beta-lactamase.
Mol.Microbiol., 60, 2006
|
|
2WH9
 
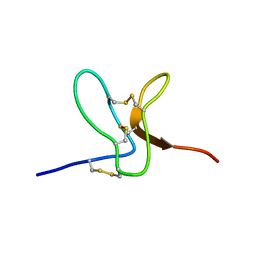 | | Solution structure of GxTX-1E | | Descriptor: | GUANGXITOXIN-1EGXTX-1E | | Authors: | Lee, S.K, Jung, H.H, Lee, J.Y, Lee, C.W, Kim, J.I. | | Deposit date: | 2009-05-02 | | Release date: | 2010-05-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Gxtx-1E, a High Affinity Tarantula Toxin Interacting with Voltage Sensors in Kv2.1 Potassium Channels.
Biochemistry, 49, 2010
|
|
3BP3
 
 | | Crystal structure of EIIB | | Descriptor: | Glucose-specific phosphotransferase enzyme IIB component, SULFATE ION | | Authors: | Cha, S.S, Jung, H.I, An, Y.J. | | Deposit date: | 2007-12-18 | | Release date: | 2008-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Analyses of Mlc-IIBGlc interaction and a plausible molecular mechanism of Mlc inactivation by membrane sequestration.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BP8
 
 | | Crystal structure of Mlc/EIIB complex | | Descriptor: | ACETATE ION, PTS system glucose-specific EIICB component, Putative NAGC-like transcriptional regulator, ... | | Authors: | An, Y.J, Jung, H.I, Cha, S.S. | | Deposit date: | 2007-12-18 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Analyses of Mlc-IIBGlc interaction and a plausible molecular mechanism of Mlc inactivation by membrane sequestration
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6MQ8
 
 | | Binary structure of DNA polymerase eta in complex with templating hypoxanthine | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*IP*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Hawkins, M.A, Jung, H, Lee, S. | | Deposit date: | 2018-10-09 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structural insights into the bypass of the major deaminated purines by translesion synthesis DNA polymerase.
Biochem.J., 2020
|
|
4HN7
 
 | |
2KET
 
 | |
2KM9
 
 | | Omega conotoxin-FVIA | | Descriptor: | omega_conotoxin-FVIA | | Authors: | Lee, S, Kim, J, Lee, J, Jung, H. | | Deposit date: | 2009-07-25 | | Release date: | 2010-07-28 | | Last modified: | 2011-09-28 | | Method: | SOLUTION NMR | | Cite: | Structure of omega conotoxin-FVIA
To be Published
|
|
6MP3
 
 | |
2KDU
 
 | | Structural basis of the Munc13-1/Ca2+-Calmodulin interaction: A novel 1-26 calmodulin binding motif with a bipartite binding mode | | Descriptor: | CALCIUM ION, Calmodulin, Protein unc-13 homolog A | | Authors: | Rodriguez-Castaneda, F.A, Maestre-Martinez, M, Coudevylle, N, Dimova, K, Jahn, O, Junge, H, Becker, S, Brose, N, Carlomagno, T, Griesinger, C. | | Deposit date: | 2009-01-19 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Modular architecture of Munc13/calmodulin complexes: dual regulation by Ca2+ and possible function in short-term synaptic plasticity.
Embo J., 29, 2010
|
|
1S7R
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Kb in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
1S7S
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Kb in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
1S7Q
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Kb in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
1S7X
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
