7U7N
 
 | | IL-27 quaternary receptor signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-27 receptor subunit alpha, ... | | Authors: | Caveney, N.A, Glassman, C.R, Jude, K.M, Tsutsumi, N, Garcia, K.C. | | Deposit date: | 2022-03-07 | | Release date: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of the IL-27 quaternary receptor signaling complex.
Elife, 11, 2022
|
|
7N2O
 
 | | AS4.2-YEIH-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2Q
 
 | | AS4.3-YEIH-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AS4.3 T cell receptor alpha chain, AS4.3 T cell receptor beta chain, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2S
 
 | | AS3.1-PRPF3-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2R
 
 | | AS4.3-PRPF3-HLA*B27 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, AS4.3 T cell receptor alpha chain, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2N
 
 | | TCR-antigen complex AS4.2-PRPF3-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2P
 
 | | AS4.3-RNASEH2b-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AS4.3 T cell receptor alpha chain, AS4.3 T cell receptor beta chain, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
8VSP
 
 | | Cryo-EM structure of human invariant chain in complex with HLA-DQ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, ... | | Authors: | Wang, N, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural insights into human MHC-II association with invariant chain.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VRW
 
 | | Cryo-EM structure of human invariant chain in complex with HLA-DR15 | | Descriptor: | HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Wang, N, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2024-01-22 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights into human MHC-II association with invariant chain.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5T5W
 
 | | Structure of an affinity matured lambda-IFN/IFN-lambdaR1/IL-10Rbeta receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon lambda receptor 1, Interferon lambda-3, ... | | Authors: | Mendoza, J.L, Jude, K.M, Garcia, K.C. | | Deposit date: | 2016-08-31 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | The IFN-lambda-IFN-lambda R1-IL-10R beta Complex Reveals Structural Features Underlying Type III IFN Functional Plasticity.
Immunity, 46, 2017
|
|
3E2L
 
 | | Crystal Structure of the KPC-2 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) | | Descriptor: | Beta-lactamase inhibitory protein, Carbapenemase | | Authors: | Hanes, M.S, Jude, K.M, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2008-08-05 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and biochemical characterization of the interaction between KPC-2 beta-lactamase and beta-lactamase inhibitor protein
Biochemistry, 48, 2009
|
|
3E2K
 
 | | Crystal Structure of the KPC-2 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) | | Descriptor: | Beta-lactamase inhibitory protein, Carbapenemase | | Authors: | Hanes, M.S, Jude, K.M, Berger, J.M, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2008-08-05 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical characterization of the interaction between KPC-2 beta-lactamase and beta-lactamase inhibitor protein
Biochemistry, 48, 2009
|
|
6BGA
 
 | | 2B4 I-Ek TCR-MHC complex with affinity-enhancing Velcro peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 peptide,MHC I-Ek B chain, ... | | Authors: | Gee, M.H, Sibener, L.V, Birnbaum, M.E, Jude, K.M, Garcia, K.C. | | Deposit date: | 2017-10-27 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Stress-testing the relationship between T cell receptor/peptide-MHC affinity and cross-reactivity using peptide velcro.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2NN7
 
 | | Structure of inhibitor binding to Carbonic Anhydrase I | | Descriptor: | Carbonic anhydrase 1, DIMETHYL SULFOXIDE, ETHYL 3-[4-(AMINOSULFONYL)PHENYL]PROPANOATE, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
2NNG
 
 | | Structure of inhibitor binding to Carbonic Anhydrase II | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONAMIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
2NNS
 
 | |
2NNV
 
 | | Structure of inhibitor binding to Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase 2, ETHYL 3-[4-(AMINOSULFONYL)PHENYL]PROPANOATE, GLYCEROL, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
2NMX
 
 | | Structure of inhibitor binding to Carbonic Anhydrase I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 1, N-{2-[4-(AMINOSULFONYL)PHENYL]ETHYL}ACETAMIDE, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II
J.Am.Chem.Soc., 129, 2007
|
|
2NN1
 
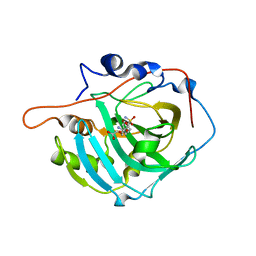 | | Structure of inhibitor binding to Carbonic Anhydrase I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[4-(AMINOSULFONYL)PHENYL]PROPANOIC ACID, Carbonic anhydrase 1, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II
J.Am.Chem.Soc., 129, 2007
|
|
2NNO
 
 | | Structure of inhibitor binding to Carbonic Anhydrase II | | Descriptor: | 3-[4-(AMINOSULFONYL)PHENYL]PROPANOIC ACID, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
7K7R
 
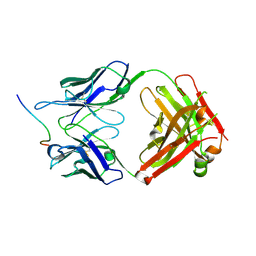 | | EBNA1 peptide AA386-405 with Fab MS39p2w174 | | Descriptor: | CHLORIDE ION, EBNA1 peptide AA386-405, Fab HC MS39p2w174, ... | | Authors: | Lanz, T.V, Robinson, W.H, Jude, K.M. | | Deposit date: | 2020-09-23 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Clonally expanded B cells in multiple sclerosis bind EBV EBNA1 and GlialCAM.
Nature, 603, 2022
|
|
6NDZ
 
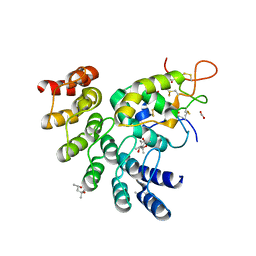 | | Designed repeat protein in complex with Fz8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ACETIC ACID, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.264 Å) | | Cite: | Receptor subtype discrimination using extensive shape complementary designed interfaces.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NE2
 
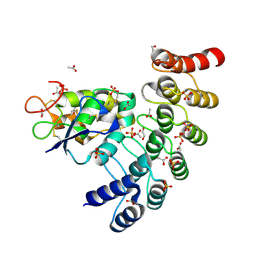 | | Designed repeat protein in complex with Fz7 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2018-12-15 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Receptor subtype discrimination using extensive shape complementary designed interfaces.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NE1
 
 | | Designed repeat protein in complex with Fz4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4, Pfs, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2018-12-15 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Receptor subtype discrimination using extensive shape complementary designed interfaces.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NE4
 
 | | Designed repeat protein specifically in complex with Fz7CRD | | Descriptor: | 1,2-ETHANEDIOL, Designed repeat binding protein, Frizzled-7, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2018-12-16 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Receptor subtype discrimination using extensive shape complementary designed interfaces.
Nat.Struct.Mol.Biol., 26, 2019
|
|
