6ZVY
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H-P174R LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineered HaloTag variants for fluorescence lifetime multiplexing.
Nat.Methods, 19, 2022
|
|
6ZVV
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174W LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174W LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE
To Be Published
|
|
6Y7B
 
 | | X-ray structure of the Haloalkane dehalogenase HaloTag7 labeled with a chloroalkane-carbopyronine fluorophore substrate | | Descriptor: | 4-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]-2-[3-(dimethylamino)-6-(dimethyl-$l^{4}-azanylidene)-10,10-dimethyl-anthracen-9-yl]benzoic acid, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-02-28 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
4TRT
 
 | |
5TVV
 
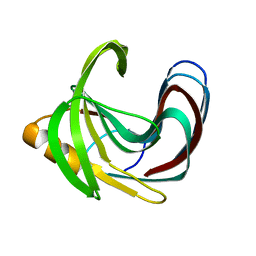 | | Computationally Designed Fentanyl Binder - Fen49* Apo | | Descriptor: | Endo-1,4-beta-xylanase A, POTASSIUM ION | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, A.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-10 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
8VHH
 
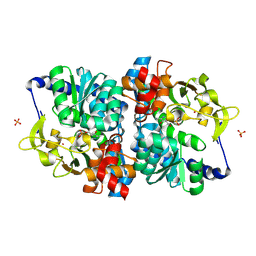 | | Engineered holo tryptophan synthase (Tm9D8*) derived from T. maritima TrpB | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Porter, N.J, Johnston, K.E, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2024-01-02 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A combinatorially complete epistatic fitness landscape in an enzyme active site.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5TVY
 
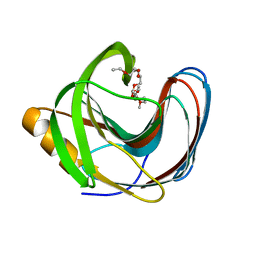 | | Computationally Designed Fentanyl Binder - Fen49 | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Endo-1,4-beta-xylanase A | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, M.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-10 | | Release date: | 2017-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
8OVN
 
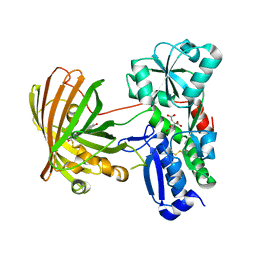 | | X-ray structure of the SF-iGluSnFR-S72A | | Descriptor: | CITRIC ACID, Putative periplasmic binding transport protein,Green fluorescent protein | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the SF-iGluSnFR-S72A
To Be Published
|
|
8OVO
 
 | | X-ray structure of the SF-iGluSnFR-S72A in complex with L-aspartate | | Descriptor: | ASPARTIC ACID, Putative periplasmic binding transport protein,Green fluorescent protein | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the SF-iGluSnFR-S72A in complex with L-aspartate
To Be Published
|
|
6Y7A
 
 | | X-ray structure of the Haloalkane dehalogenase HaloTag7 labeled with a chloroalkane-tetramethylrhodamine fluorophore substrate | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-02-28 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
6Y8P
 
 | | Crystal structure of SNAP-tag labeled with a benzyl-tetramethylrhodamine fluorophore | | Descriptor: | 1,2-ETHANEDIOL, O6-alkylguanine-DNA alkyltransferase mutant, ZINC ION, ... | | Authors: | Gotthard, G, Tanzer, T, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-03-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
5WBF
 
 | | Double CACHE (dCACHE) sensing domain of TlpC chemoreceptor from Helicobacter pylori | | Descriptor: | GLYCEROL, LACTIC ACID, Methyl-accepting chemotaxis transducer (TlpC) | | Authors: | Machuca, M.A, Johnson, K.S, Liu, Y.C, Steer, D.L, Ottemann, K.M, Roujeinikova, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Helicobacter pylori chemoreceptor TlpC mediates chemotaxis to lactate.
Sci Rep, 7, 2017
|
|
7ZJ0
 
 | | X-ray structure of the haloalkane dehalogenase HaloTag7 bound to a pentylmethanesulfonamide tetramethylrhodamine ligand (TMR-S5) | | Descriptor: | GLYCEROL, Haloalkane dehalogenase, [9-[2-carboxy-5-[2-[2-[5-(methylsulfonylamino)pentoxy]ethoxy]ethylcarbamoyl]phenyl]-6-(dimethylamino)xanthen-3-ylidene]-dimethyl-azanium | | Authors: | Tarnawski, M, Kompa, J, Johnsson, K, Hiblot, J. | | Deposit date: | 2022-04-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exchangeable HaloTag Ligands for Super-Resolution Fluorescence Microscopy.
J.Am.Chem.Soc., 145, 2023
|
|
4HWK
 
 | | Crystal structure of human sepiapterin reductase in complex with sulfapyridine | | Descriptor: | 4-amino-N-(pyridin-2-yl)benzenesulfonamide, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Groenlund Pedersen, M, Pojer, F, Johnsson, K. | | Deposit date: | 2012-11-08 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tetrahydrobiopterin biosynthesis as an off-target of sulfa drugs.
Science, 340, 2013
|
|
3FFE
 
 | | Structure of Achromobactin Synthetase Protein D, (AcsD) | | Descriptor: | AcsD | | Authors: | McMahon, S.A, Liu, H, Carter, L, Oke, M, Johnson, K.A, Schmelz, S, Challis, G.L, White, M.F, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2008-12-03 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
6ZVU
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE
To Be Published
|
|
6ZVX
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE
To Be Published
|
|
6ZVW
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE
To Be Published
|
|
3C6W
 
 | | Crystal structure of the ING5 PHD finger in complex with H3K4me3 peptide | | Descriptor: | H3K4me3 histone peptide, Inhibitor of growth protein 5, ZINC ION | | Authors: | Champagne, K.S, Pena, P.V, Johnson, K, Kutateladze, T.G. | | Deposit date: | 2008-02-05 | | Release date: | 2008-06-03 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the ING5 PHD finger in complex with an H3K4me3 histone peptide.
Proteins, 72, 2008
|
|
4J7U
 
 | | Crystal structure of human sepiapterin reductase in complex with sulfathiazole | | Descriptor: | 4-amino-N-(1,3-thiazol-2-yl)benzenesulfonamide, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Groenlund Pedersen, M, Pojer, F, Johnsson, K. | | Deposit date: | 2013-02-14 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Tetrahydrobiopterin biosynthesis as an off-target of sulfa drugs.
Science, 340, 2013
|
|
1GJY
 
 | | The X-ray structure of the Sorcin Calcium Binding Domain (SCBD) provides insight into the phosphorylation and calcium dependent processess | | Descriptor: | SORCIN, SULFATE ION | | Authors: | Ilari, A, Johnson, K.A, Nastopoulos, V, Tsernoglou, D, Chiancone, E. | | Deposit date: | 2001-08-06 | | Release date: | 2002-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Sorcin Calcium Binding Domain Provides a Model of Ca(2+)-Dependent Processes in the Full-Length Protein
J.Mol.Biol., 317, 2002
|
|
2W6W
 
 | | Crystal structure of recombinant Sperm Whale Myoglobin under 1atm of Xenon | | Descriptor: | GLYCEROL, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miele, A.E, Draghi, F, Renzi, F, Sciara, G, Johnson, K.A, Vallone, B, Brunori, M, Savino, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Pattern of Cavities in Globins: The Case of Human Hemoglobin.
Biopolymers, 91, 2009
|
|
2W72
 
 | | DEOXYGENATED STRUCTURE OF A DISTAL SITE HEMOGLOBIN MUTANT PLUS XE | | Descriptor: | HUMAN HEMOGLOBIN A, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Miele, A.E, Draghi, F, Sciara, G, Johnson, K.A, Renzi, F, Vallone, B, Brunori, M, Savino, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Pattern of Cavities in Globins: The Case of Human Hemoglobin.
Biopolymers, 91, 2009
|
|
1DK4
 
 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE | | Descriptor: | INOSITOL MONOPHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Stec, B, Yang, H, Johnson, K.A, Chen, L, Roberts, M.F. | | Deposit date: | 1999-12-06 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MJ0109 is an enzyme that is both an inositol monophosphatase and the 'missing' archaeal fructose-1,6-bisphosphatase.
Nat.Struct.Biol., 7, 2000
|
|
1DUX
 
 | | ELK-1/DNA STRUCTURE REVEALS HOW RESIDUES DISTAL FROM DNA-BINDING SURFACE AFFECT DNA-RECOGNITION | | Descriptor: | DNA (5'-D(*AP*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP*T)-3'), ETS-DOMAIN PROTEIN ELK-1 | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 2000-01-19 | | Release date: | 2000-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the elk-1-DNA complex reveals how DNA-distal residues affect ETS domain recognition of DNA.
Nat.Struct.Biol., 7, 2000
|
|
