6QZ1
 
 | | Structure of MHETase from Ideonella sakaiensis | | Descriptor: | BENZOIC ACID, CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase | | Authors: | Allen, M.D, Johnson, C.W, Knott, B.C, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2019-03-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and engineering of a two-enzyme system for plastics depolymerization.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3UMS
 
 | | Crystal structure of the G202A mutant of human G-alpha-i1 | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lambert, N.A, Johnston, C.A, Cappell, S.D, Kuravi, S, Kimple, A.J, Willard, F.S, Siderovski, D.P. | | Deposit date: | 2011-11-14 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Correction for Regulators of G-protein Signaling accelerate GPCR signaling kinetics and govern sensitivity solely by accelerating GTPase activity
Proc.Natl.Acad.Sci.USA, 2012
|
|
2GBL
 
 | | Crystal Structure of Full Length Circadian Clock Protein KaiC with Phosphorylation Sites | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Williams, D.R, Pattanayek, S, Xu, Y, Mori, T, Johnson, C.H, Stewart, P.L, Egli, M. | | Deposit date: | 2006-03-10 | | Release date: | 2007-01-23 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of KaiA-KaiC protein interactions in the cyano-bacterial circadian clock using hybrid structural methods.
Embo J., 25, 2006
|
|
1W4J
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | PYRUVATE DEHYDROGENASE E2 | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-fast barrier-limited folding in the peripheral subunit-binding domain family.
J. Mol. Biol., 353, 2005
|
|
1W4G
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state folding transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4E
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4K
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | PYRUVATE DEHYDROGENASE E2 | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4I
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | PYRUVATE DEHYDROGENASE E2 | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
8ELS
 
 | | HRAS R97A Crystal Form 1 R-state | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Davis, D, Johnson, C.W, Mattos, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.267 Å) | | Cite: | Allosteric site variants affect GTP hydrolysis on Ras.
Protein Sci., 32, 2023
|
|
8FG4
 
 | | HRAS R97L Crystal Form 1 | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Fetics, S, Johnson, C.W, Mattos, C. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Allosteric site variants affect GTP hydrolysis on Ras.
Protein Sci., 32, 2023
|
|
8FG3
 
 | | HRAS R97L Crystal Form 2 | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Fetics, S, Johnson, C.W, Mattos, C. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Allosteric site variants affect GTP hydrolysis on Ras.
Protein Sci., 32, 2023
|
|
8ELW
 
 | | HRAS R97A Crystal Form 1 T-State | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Davis, K, Johnson, C.W, Mattos, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Allosteric site variants affect GTP hydrolysis on Ras.
Protein Sci., 32, 2023
|
|
6QZ2
 
 | | Structure of MHETase from Ideonella sakaiensis | | Descriptor: | CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase | | Authors: | Allen, M.D, Johnson, C.W, Knott, B.C, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2019-03-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and engineering of a two-enzyme system for plastics depolymerization.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6QZ3
 
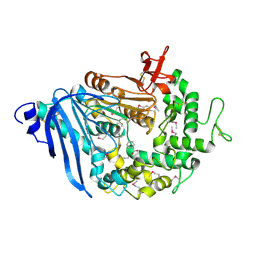 | | Structure of MHETase from Ideonella sakaiensis | | Descriptor: | BENZOIC ACID, CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase | | Authors: | Allen, M.D, Johnson, C.W, Knott, B.C, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2019-03-11 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization and engineering of a two-enzyme system for plastics depolymerization.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NP0
 
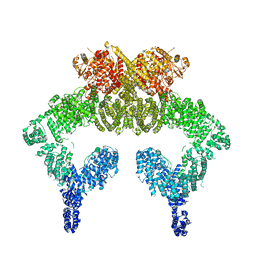 | | Closed dimer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
5NP1
 
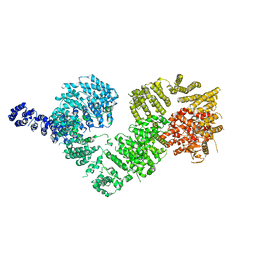 | | Open protomer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
6QZ4
 
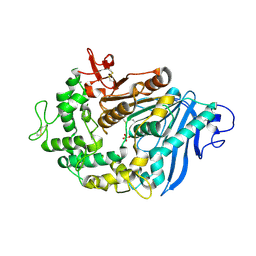 | | Structure of MHETase from Ideonella sakaiensis | | Descriptor: | CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase, SULFATE ION | | Authors: | Allen, M.D, Johnson, C.W, Knott, B.C, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2019-03-11 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and engineering of a two-enzyme system for plastics depolymerization.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5N2W
 
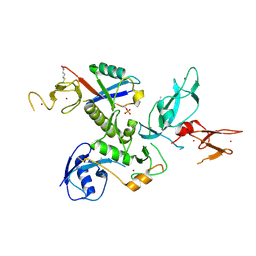 | | WT-Parkin and pUB complex | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, Polyubiquitin-B, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5N38
 
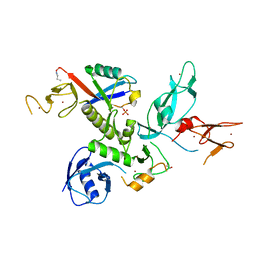 | | S65DParkin and pUB complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5KC1
 
 | | Structure of the C-terminal dimerization domain of Atg38 | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Autophagy-related protein 38, ... | | Authors: | Ohashi, Y, Soler, N, Garcia-Ortegon, M, Zhang, L, Perisic, O, Masson, G.R, Johnson, C.M, Williams, R.J. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
6EQH
 
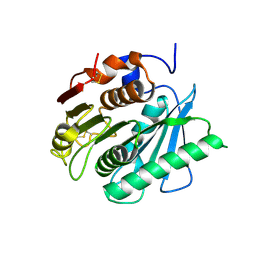 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis in spacegroup C2221 | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EQD
 
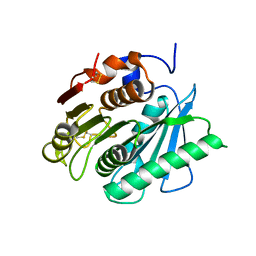 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis collected at long wavelength | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EQG
 
 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis in spacegroup P21 | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EQE
 
 | | High resolution crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SODIUM ION | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5UHV
 
 | | wild-type NRas bound to GppNHp | | Descriptor: | GLYCEROL, GTPase NRas, MAGNESIUM ION, ... | | Authors: | Reid, D, Johnson, C, Salter, S, Mattos, C. | | Deposit date: | 2017-01-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.672 Å) | | Cite: | The small GTPases K-Ras, N-Ras, and H-Ras have distinct biochemical properties determined by allosteric effects.
J. Biol. Chem., 292, 2017
|
|
