6A2V
 
 | | Crystal structure of Hcp protein | | Descriptor: | Type VI secretion system tube protein Hcp | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2018-06-13 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.588 Å) | | Cite: | Structural basis for the pathogenesis of Campylobacter jejuni Hcp1, a structural and effector protein of the Type VI Secretion System.
FEBS J., 285, 2018
|
|
7EDD
 
 | |
7FCF
 
 | | Crystal structure of T6SS Hcp protein | | Descriptor: | Fimbrial protein | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bacterial antagonism of Chromobacterium haemolyticum and characterization of its putative type VI secretion system.
Res.Microbiol., 173, 2022
|
|
7CBK
 
 | | Structure of Human Neutrophil Elastase Ecotin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ecotin, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Inhibition Mechanism of Ecotin against Neutrophil Elastase by Targeting the Active Site and Secondary Binding Site.
Biochemistry, 59, 2020
|
|
5XWR
 
 | | Crystal Structure of RBBP4-peptide complex | | Descriptor: | Histone-binding protein RBBP4, MET-SER-ARG-ARG-LYS-GLN-ALA-LYS-PRO-GLN-HIS-ILE | | Authors: | Jobichen, C, Lui, B.H, Daniel, G.T, Sivaraman, J. | | Deposit date: | 2017-06-30 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XXZ
 
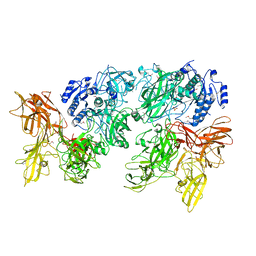 | |
5XYA
 
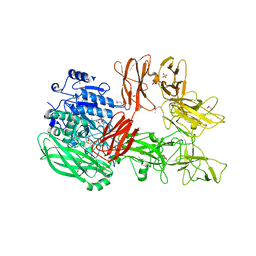 | | Crystal structure of a serine protease from Streptococcus species | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CALCIUM ION, Chemokine protease C, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2017-07-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of ScpC, a virulence protease fromStreptococcus pyogenes, reveals the functional domains and maturation mechanism.
Biochem. J., 475, 2018
|
|
5XYR
 
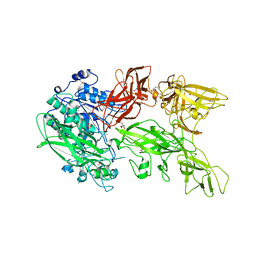 | | Crystal structure of a serine protease from Streptococcus species | | Descriptor: | CALCIUM ION, CHLORIDE ION, Chemokine protease C, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2017-07-10 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of ScpC, a virulence protease fromStreptococcus pyogenes, reveals the functional domains and maturation mechanism.
Biochem. J., 475, 2018
|
|
7DEY
 
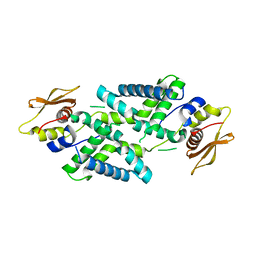 | | Structure of Dicer from Pichia stipitis | | Descriptor: | RNase III | | Authors: | Jobichen, C, Jingru, C. | | Deposit date: | 2020-11-05 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Structural and mechanistic insight into stem-loop RNA processing by yeast Pichia stipitis Dicer.
Protein Sci., 30, 2021
|
|
4NQF
 
 | |
4KT5
 
 | |
5E38
 
 | | Structural basis of mapping the spontaneous mutations with 5-flourouracil in uracil phosphoribosyltransferase from Mycobacterium tuberculosis | | Descriptor: | Uracil phosphoribosyltransferase | | Authors: | Ghode, P, Jobichen, C, Ramachandran, S, Bifani, P, Sivaraman, J. | | Deposit date: | 2015-10-02 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of mapping the spontaneous mutations with 5-flurouracil in uracil phosphoribosyltransferase from Mycobacterium tuberculosis
Biochem.Biophys.Res.Commun., 467, 2015
|
|
6LKB
 
 | | Crystal Structure of the peptidylprolyl isomerase domain of Arabidopsis thaliana CYP71. | | Descriptor: | COBALT (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lakhanpal, S, Jobichen, C, Swaminathan, K. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and functional analyses of the PPIase domain of Arabidopsis thaliana CYP71 reveal its catalytic activity toward histone H3.
Febs Lett., 595, 2021
|
|
6KOR
 
 | | Crystal structure of the RRM domain of SYNCRIP | | Descriptor: | Heterogeneous nuclear ribonucleoprotein Q | | Authors: | Chen, Y, Chan, J, Chen, W, Jobichen, C. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | SYNCRIP, a new player in pri-let-7a processing.
Rna, 26, 2020
|
|
8GWR
 
 | | Near full length Kidney type Glutaminase in complex with 2,2-Dimethyl-2,3-Dihydrobenzo[a] Phenanthridin-4(1H)-one (DDP) | | Descriptor: | 2,2-dimethyl-1,3-dihydrobenzo[a]phenanthridin-4-one, Glutaminase kidney isoform, mitochondrial | | Authors: | Shankar, S, Jobichen, C, Sivaraman, J. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | A novel allosteric site employs a conserved inhibition mechanism in human kidney-type glutaminase.
Febs J., 290, 2023
|
|
3TVD
 
 | | Crystal Structure of Mouse RhoA-GTP complex | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Swaminathan, K, Pal, K, Jobichen, C. | | Deposit date: | 2011-09-20 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Crystal structure of mouse RhoA:GTPgammaS complex in a centered lattice.
J.Struct.Funct.Genom., 13, 2012
|
|
8H9D
 
 | | Crystal structure of Cas12a protein | | Descriptor: | Cas12A, MAGNESIUM ION, RNA (5'-R(P*AP*AP*UP*UP*UP*CP*UP*AP*CP*UP*AP*AP*GP*UP*GP*UP*AP*GP*AP*UP*C)-3'), ... | | Authors: | Jianwei, L, Jobichen, C, Sivaraman, J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of apo Cas12a and its complex with crRNA and DNA reveal the dynamics of ternary complex formation and target DNA cleavage.
Plos Biol., 21, 2023
|
|
3WXX
 
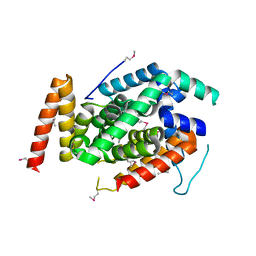 | | Crystal Structure of a T3SS complex from Aeromonas hydrophila | | Descriptor: | AcrH, AopB, MAGNESIUM ION | | Authors: | Nguyen, V.S, Jobichen, C, Sivaraman, J, Henry, Y.K.M. | | Deposit date: | 2014-08-12 | | Release date: | 2015-10-14 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of AcrH-AopB Chaperone-Translocator Complex Reveals a Role for Membrane Hairpins in Type III Secretion System Translocon Assembly
Structure, 23, 2015
|
|
5BN5
 
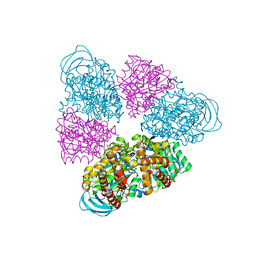 | | Structural basis for a unique ATP synthase core complex from Nanoarcheaum equitans | | Descriptor: | NEQ263, SULFATE ION, V-type ATP synthase alpha chain | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
5BN4
 
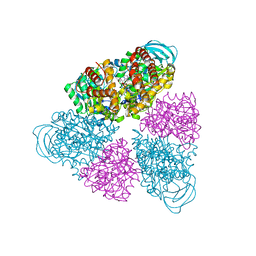 | | Structure of a unique ATP synthase NeqA-NeqB in complex with ANP from Nanoarcheaum equitans | | Descriptor: | MAGNESIUM ION, NEQ263, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
5BN3
 
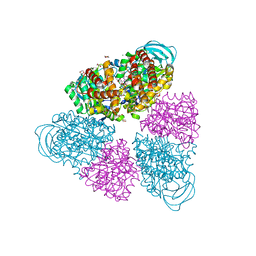 | | Structure of a unique ATP synthase NeqA-NeqB in complex with ADP from Nanoarcheaum equitans | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
5BO5
 
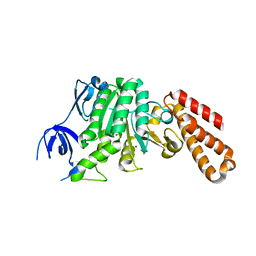 | | Structure of a unique ATP synthase subunit NeqB from Nanoarcheaum equitans | | Descriptor: | MAGNESIUM ION, NEQ263, SULFATE ION | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
7E50
 
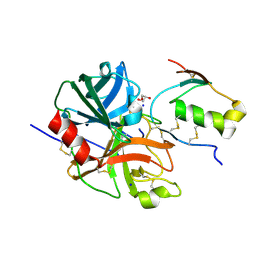 | | Crystal structure of human microplasmin in complex with kazal-type inhibitor AaTI | | Descriptor: | AAEL006007-PA, GLYCEROL, Plasminogen, ... | | Authors: | Varsha, A.W, Jobichen, C, Mok, Y.K. | | Deposit date: | 2021-02-16 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Aedes aegypti trypsin inhibitor in complex with mu-plasmin reveals role for scaffold stability in Kazal-type serine protease inhibitor.
Protein Sci., 31, 2022
|
|
7EQZ
 
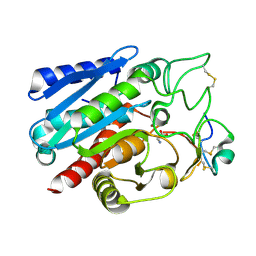 | | Crystal structure of Carboxypeptidase B complexed with Potato Carboxypeptidase Inhibitor | | Descriptor: | Carboxypeptidase B, GLYCINE, Metallocarboxypeptidase inhibitor, ... | | Authors: | Choong, Y.K, Gavor, E, Jobichen, C, Sivaraman, J. | | Deposit date: | 2021-05-05 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Aedes aegypti carboxypeptidase B1-inhibitor complex uncover the disparity between mosquito and non-mosquito insect carboxypeptidase inhibition mechanism.
Protein Sci., 30, 2021
|
|
7E4U
 
 | |
