5YGP
 
 | | Human TNFRSF25 death domain mutant-D412E | | Descriptor: | SULFATE ION, TNFRSF25 death domain, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yin, X, Jin, T.C. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
6J3X
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 with Maltotriose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucan 1,4-alpha-maltotetraohydrolase, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2019-01-06 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Maltotetraose-forming amylase from Pseudomonas saccharophila STB07
To Be Published
|
|
6IYG
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 with Maltotetraose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucan 1,4-alpha-maltotetraohydrolase, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2018-12-15 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Maltotetraose-forming amylase from Pseudomonas saccharophila STB07
To Be Published
|
|
6AIJ
 
 | | Cyclodextrin glycosyltransferase from Paenibacillus macerans mutant N603D | | Descriptor: | CALCIUM ION, Cyclomaltodextrin glucanotransferase | | Authors: | Li, C.M, Ban, X.F, Li, Z.F, Li, Y.L, Cheng, S.D, Zhang, C.Y, Jin, T.C, Gu, Z.B. | | Deposit date: | 2018-08-24 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Cyclodextrin glycosyltransferase from Paenibacillus macerans mutant N603D
To Be Published
|
|
8HN4
 
 | |
8IDX
 
 | | Structure of p205 HIN | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Interferon-activable protein 205-B, ... | | Authors: | Li, Y.L, Jin, T.C. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of p205 HIN
to Be published
|
|
8I6K
 
 | | Structure of hMNDA HIN with dsDNA | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Li, Y.L, Jin, T.C. | | Deposit date: | 2023-01-28 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism of dsDNA recognition by the hMNDA HIN domain: New insights into the DNA-binding model of a PYHIN protein.
Int.J.Biol.Macromol., 245, 2023
|
|
6IWK
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 | | Descriptor: | CALCIUM ION, GLYCEROL, Glucan 1,4-alpha-maltotetraohydrolase | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
7V4Q
 
 | |
5ZH6
 
 | | Crystal structure of Parvalbumin SPV-II of Mustelus griseus | | Descriptor: | CALCIUM ION, Parvalbumin SPV-II, SULFATE ION | | Authors: | Yang, R.Q, Chen, Y.L, Jin, T.C, Cao, M.J. | | Deposit date: | 2018-03-11 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Purification, Characterization, and Crystal Structure of Parvalbumins, the Major Allergens in Mustelus griseus.
J. Agric. Food Chem., 66, 2018
|
|
7ESV
 
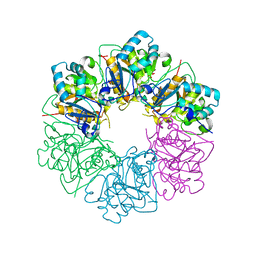 | |
7ESQ
 
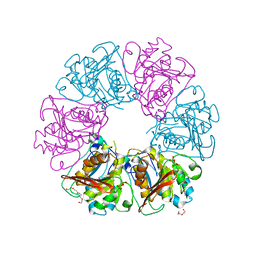 | |
7ESP
 
 | |
7ESO
 
 | |
5ZHQ
 
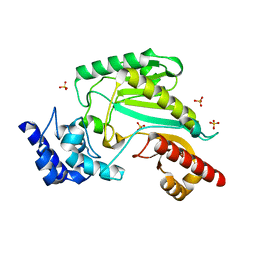 | |
5ZGM
 
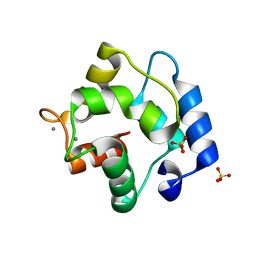 | | Crystal Structure of Parvalbumin SPVI, the Major Allergens in Mustelus griseus | | Descriptor: | CALCIUM ION, Parvalbumin SPVI, SULFATE ION | | Authors: | Yang, R.Q, Chen, Y.L, Jin, T.C, Cao, M.J. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Purification, Characterization, and Crystal Structure of Parvalbumins, the Major Allergens in Mustelus griseus.
J. Agric. Food Chem., 66, 2018
|
|
7VOA
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with aRBD5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpaca nanobody | | Authors: | Ma, H, Zeng, W.H, Jin, T.C. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hetero-bivalent nanobodies provide broad-spectrum protection against SARS-CoV-2 variants of concern including Omicron.
Cell Res., 32, 2022
|
|
6KI0
 
 | | Crystal Structure of Human ASC-CARD | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Xu, Z.H, Jin, T.C. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homotypic CARD-CARD interaction is critical for the activation of NLRP1 inflammasome.
Cell Death Dis, 12, 2021
|
|
6AG0
 
 | | The X-ray Crystallographic Structure of Maltooligosaccharide-forming Amylase from Bacillus stearothermophilus STB04 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase, CALCIUM ION | | Authors: | Li, Z.F, Li, Y.L, Ban, X.F, Zhang, C.Y, Jin, T.C, Xie, X.F, Gu, Z.B, Li, C.M. | | Deposit date: | 2018-08-09 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04.
Int.J.Biol.Macromol., 138, 2019
|
|
6KXG
 
 | | Crystal structure of caspase-11-CARD | | Descriptor: | caspase-11-CARD | | Authors: | Liu, M.Z.Y, Jin, T.C. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Crystal structure of caspase-11 CARD provides insights into caspase-11 activation.
Cell Discov, 6, 2020
|
|
6JQB
 
 | | The structure of maltooligosaccharide-forming amylase from Pseudomonas saccharophila STB07 with pseudo-maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, ACARBOSE DERIVED HEPTASACCHARIDE, CALCIUM ION, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2019-03-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
7V4R
 
 | | The crystal structure of KFDV NS3H bound with Pi | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, Serine protease NS3 | | Authors: | Zhang, C.Y, Jin, T.C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kyasanur Forest disease virus NS3 helicase: Insights into structure, activity, and inhibitors.
Int.J.Biol.Macromol., 2023
|
|
6JOY
 
 | | The X-ray Crystallographic Structure of Branching Enzyme from Rhodothermus obamensis STB05 | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB | | Authors: | Li, Z.F, Ban, X.F, Jiang, H.M, Wang, Z, Jin, T.C, Li, C.M, Gu, Z.B. | | Deposit date: | 2019-03-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Flexible Loop in Carbohydrate-Binding Module 48 Allosterically Modulates Substrate Binding of the 1,4-alpha-Glucan Branching Enzyme.
J.Agric.Food Chem., 69, 2021
|
|
