8HOP
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Nap-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Nap-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HOQ
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Phe(4CF3)-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Phe(4CF3)-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HON
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Tyr-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Tyr-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5ZQ0
 
 | | Crystal structure of spRlmCD with U747loop RNA | | Descriptor: | RNA (5'-R(*GP*UP*(MUM)P*GP*AP*AP*AP*A)-3'), S-ADENOSYL-L-HOMOCYSTEINE, Uncharacterized RNA methyltransferase SP_1029 | | Authors: | Jiang, Y.Y, Yu, H.L. | | Deposit date: | 2018-04-17 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unveiling the structural features that determine the dual methyltransferase activities of Streptococcus pneumoniae RlmCD
PLoS Pathog., 14, 2018
|
|
5ZQ8
 
 | | Crystal structure of spRlmCD with U747 stemloop RNA | | Descriptor: | NICKEL (II) ION, RNA (5'-R(*CP*CP*GP*UP*(MUM)P*GP*AP*AP*AP*AP*GP*G)-3'), S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Jiang, Y.Y, Yu, H.L. | | Deposit date: | 2018-04-17 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Unveiling the structural features that determine the dual methyltransferase activities of Streptococcus pneumoniae RlmCD
PLoS Pathog., 14, 2018
|
|
6A4M
 
 | | Structure of urate oxidase from Bacillus subtilis 168 | | Descriptor: | GLYCEROL, Uric acid degradation bifunctional protein PucL | | Authors: | Jiang, Y.Y. | | Deposit date: | 2018-06-20 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Urate Oxidase from Bacillus Subtilis 168
Crystallography Reports, 64, 2019
|
|
3K04
 
 | | Crystal Structure of CNG mimicking NaK mutant, NaK-DTPP, Na+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein NaK, SODIUM ION | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5ZTH
 
 | | Crystal structure of spRlmCD with U1939loop RNA at 3.24 angstrom | | Descriptor: | RNA (5'-R(P*AP*AP*AP*(MUM)P*UP*CP*CP*U)-3'), S-ADENOSYL-L-HOMOCYSTEINE, Uncharacterized RNA methyltransferase SP_1029 | | Authors: | Jiang, Y.Y, Yu, H.L. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Unveiling the structural features that determine the dual methyltransferase activities of Streptococcus pneumoniae RlmCD
PLoS Pathog., 14, 2018
|
|
7WDH
 
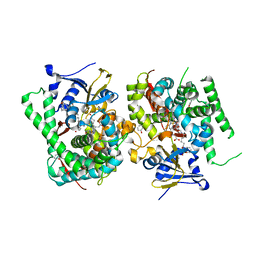 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine, phenol and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Engineering Cytochrome P450BM3 Enzymes for Direct Nitration of Unsaturated Hydrocarbons.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WDG
 
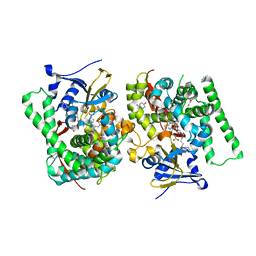 | | Crystal structure of the P450 BM3 heme domain mutant F87L in complex with N-imidazolyl-hexanoyl-L-phenylalanine, phenol and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Engineering Cytochrome P450BM3 Enzymes for Direct Nitration of Unsaturated Hydrocarbons.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3K0D
 
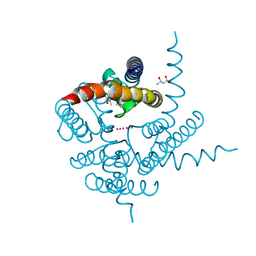 | | Crystal Structure of CNG mimicking NaK mutant, NaK-ETPP, K+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein NaK | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3K08
 
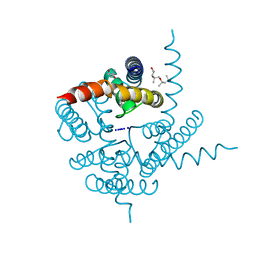 | | Crystal Structure of CNG mimicking NaK mutant, NaK-NTPP, Na+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein NaK, SODIUM ION | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3K06
 
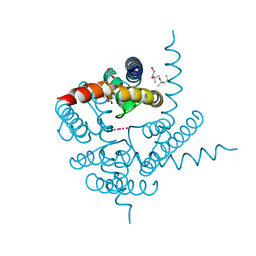 | | Crystal Structure of CNG mimicking NaK mutant, NaK-NTPP, K+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein NaK | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3K0G
 
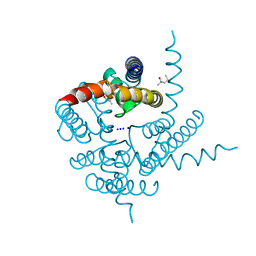 | | Crystal Structure of CNG mimicking NaK mutant, NaK-ETPP, Na+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein NaK, SODIUM ION | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8HOT
 
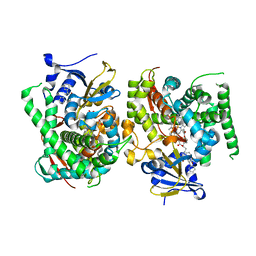 | |
8HOO
 
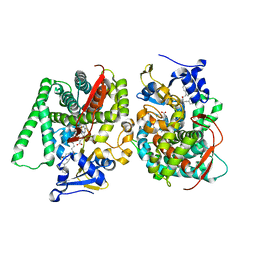 | |
5XJ2
 
 | | Structure of spRlmCD with U747 RNA | | Descriptor: | RNA (5'-R(*GP*GP*CP*AP*CP*GP*UP*GP*CP*U)-3'), S-ADENOSYL-L-HOMOCYSTEINE, Uncharacterized RNA methyltransferase SP_1029, ... | | Authors: | Jiang, Y, Gong, Q. | | Deposit date: | 2017-04-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural insights into substrate selectivity of ribosomal RNA methyltransferase RlmCD
PLoS ONE, 12, 2017
|
|
5XJ1
 
 | | Crystal structure of spRlmCD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Jiang, Y, Gong, Q. | | Deposit date: | 2017-04-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structural insights into substrate selectivity of ribosomal RNA methyltransferase RlmCD
PLoS ONE, 12, 2017
|
|
7EGN
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | H-Bonding Networks Dictate the Molecular Mechanism of H2O2 Activation by P450
Acs Catalysis, 11, 2021
|
|
5Y2V
 
 | | Strcutrue of the full-length CcmR complexed with 2-OG from Synechocystis PCC6803 | | Descriptor: | 2-OXOGLUTARIC ACID, PHOSPHATE ION, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Y2W
 
 | | Structure of Synechocystis PCC6803 CcmR regulatory domain in complex with 2-PG | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Cao, D.D, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7BV1
 
 | | Cryo-EM structure of the apo nsp12-nsp7-nsp8 complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
7BV2
 
 | | The nsp12-nsp7-nsp8 complex bound to the template-primer RNA and triphosphate form of Remdesivir(RTP) | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
7MHZ
 
 | |
5JDF
 
 | | Structural mechanisms of extracellular ion exchange and induced binding-site occlusion in the sodium-calcium exchanger NCX_Mj soaked with 2.5 mM Na+ and 1mM Ca2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, PENTADECANE, ... | | Authors: | Liao, J, Jiang, Y.X, Faraldo-Gomez, J.D. | | Deposit date: | 2016-04-16 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger.
Nat.Struct.Mol.Biol., 23, 2016
|
|
