5GZS
 
 | | Structure of VC protein | | Descriptor: | ARGININE, GGDEF family protein | | Authors: | Xu, M, Wang, Y.Z, Yang, X.A, Xie, W, Jiang, T. | | Deposit date: | 2016-10-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural studies of the periplasmic portion of the diguanylate cyclase CdgH from Vibrio cholerae.
Sci Rep, 7, 2017
|
|
6KBD
 
 | | fused To-MtbCsm1 with 2dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A),CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), MAGNESIUM ION | | Authors: | Li, T, Huo, Y, Jiang, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mycobacterium tuberculosis CRISPR/Cas system Csm1 holds clues to the evolutionary relationship between DNA polymerase and cyclase activity.
Int.J.Biol.Macromol., 170, 2020
|
|
6KC0
 
 | | fused To-MtbCsm1 with 2ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A),CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), MAGNESIUM ION | | Authors: | Li, T, Huo, Y, Jiang, T. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Mycobacterium tuberculosis CRISPR/Cas system Csm1 holds clues to the evolutionary relationship between DNA polymerase and cyclase activity.
Int.J.Biol.Macromol., 170, 2020
|
|
8I6P
 
 | | The cryo-EM structure of OsCyc1 tetramer state | | Descriptor: | Syn-copalyl diphosphate synthase, chloroplastic | | Authors: | Ma, X.L, Xu, H.F, Tong, Y.R, Luo, Y.F, Dong, Q.H, Jiang, T. | | Deposit date: | 2023-01-29 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional investigations of syn-copalyl diphosphate synthase from Oryza sativa.
Commun Chem, 6, 2023
|
|
8I6U
 
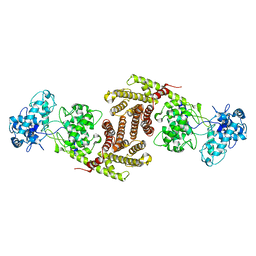 | | The cryo-EM structure of OsCyc1 dimer state | | Descriptor: | Syn-copalyl diphosphate synthase, chloroplastic | | Authors: | Ma, X.L, Xu, H.F, Tong, Y.R, Luo, Y.F, Dong, Q.H, Jiang, T. | | Deposit date: | 2023-01-29 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structural and functional investigations of syn-copalyl diphosphate synthase from Oryza sativa.
Commun Chem, 6, 2023
|
|
8I6T
 
 | | The cryo-EM structure of OsCyc1 hexamer state | | Descriptor: | Syn-copalyl diphosphate synthase, chloroplastic | | Authors: | Ma, X.L, Xu, H.F, Tong, Y.R, Luo, Y.F, Dong, Q.H, Jiang, T. | | Deposit date: | 2023-01-29 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and functional investigations of syn-copalyl diphosphate synthase from Oryza sativa.
Commun Chem, 6, 2023
|
|
8IH5
 
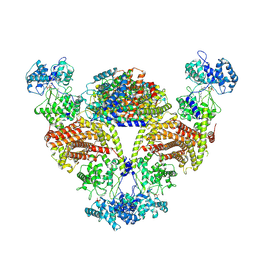 | |
2HVC
 
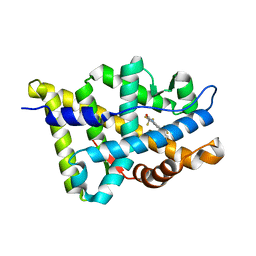 | | The Crystal Structure of Ligand-binding Domain (LBD) of human Androgen Receptor in Complex with a selective modulator LGD2226 | | Descriptor: | 6-[BIS(2,2,2-TRIFLUOROETHYL)AMINO]-4-(TRIFLUOROMETHYL)QUINOLIN-2(1H)-ONE, Androgen receptor | | Authors: | Wang, F, Liu, X.-Q, Li, H, Liang, K.-N, Miner, J.N, Hong, M, Kallel, E.A, van Oeveren, A, Zhi, L, Jiang, T. | | Deposit date: | 2006-07-28 | | Release date: | 2007-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the ligand-binding domain (LBD) of human androgen receptor in complex with a selective modulator LGD2226
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
5C70
 
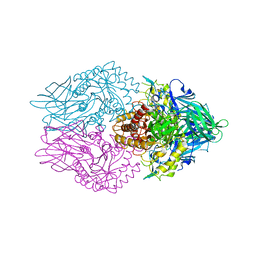 | | The structure of Aspergillus oryzae beta-glucuronidase | | Descriptor: | Glucuronidase | | Authors: | Sun, H.L, Lv, B, Huang, S, Sun, Q.F, Li, C, Jiang, T. | | Deposit date: | 2015-06-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Enhancing the Thermostability of beta-Glucuronidase by Rationally Redesigning the Catalytic Domain Based on Sequence Alignment Strategy
Ind Eng Chem Res, 55, 2016
|
|
6KFR
 
 | |
6KFU
 
 | |
6KET
 
 | |
6KFM
 
 | |
8IVQ
 
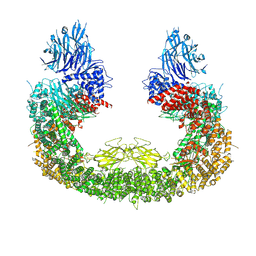 | | Cryo-EM structure of mouse BIRC6, Global map | | Descriptor: | Isoform 2 of Baculoviral IAP repeat-containing protein 6 | | Authors: | Liu, S, Jiang, T, Bu, F, Zhao, J, Wang, G, Li, N, Gao, N, Qiu, X. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanisms underlying the BIRC6-mediated regulation of apoptosis and autophagy.
Nat Commun, 15, 2024
|
|
5Y61
 
 | | YfiB-YfiR complexed with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, YfiB, YfiR | | Authors: | Zhou, L, Xu, M, Jiang, T. | | Deposit date: | 2017-08-10 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural insights into the functional role of GMP in modulating the YfiBNR system
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y62
 
 | | YfiR complexed with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, YfiR | | Authors: | Zhou, L, Xu, M, Jiang, T. | | Deposit date: | 2017-08-10 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the functional role of GMP in modulating the YfiBNR system
Biochem. Biophys. Res. Commun., 493, 2017
|
|
7CPU
 
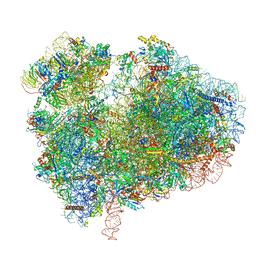 | | Cryo-EM structure of 80S ribosome from mouse kidney | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | Deposit date: | 2020-08-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
7CPV
 
 | | Cryo-EM structure of 80S ribosome from mouse testis | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | Deposit date: | 2020-08-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
