2QWN
 
 | | Crystal structure of disulfide-bond-crosslinked complex of bovine hsc70 (1-386aa)R171C and bovine Auxilin (810-910aa)D876C in the ADP*Pi state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock cognate 71 kDa protein, MAGNESIUM ION, ... | | Authors: | Jiang, J, Maes, E.G, Wang, L, Taylor, A.B, Hinck, A.P, Lafer, E.M, Sousa, R. | | Deposit date: | 2007-08-10 | | Release date: | 2007-12-18 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of J cochaperone binding and regulation of Hsp70.
Mol.Cell, 28, 2007
|
|
2QWO
 
 | | Crystal structure of disulfide-bond-crosslinked complex of bovine hsc70 (1-394aa)R171C and bovine Auxilin (810-910aa)D876C in the ADP*Pi form #1 | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Jiang, J, Maes, E.G, Wang, L, Taylor, A.B, Hinck, A.P, Lafer, E.M, Sousa, R. | | Deposit date: | 2007-08-10 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of J cochaperone binding and regulation of Hsp70.
Mol.Cell, 28, 2007
|
|
2QWM
 
 | | Crystal structure of bovine hsc70 (1-394aa)in the ADP*Vi state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Heat shock cognate 71 kDa protein, ... | | Authors: | Jiang, J, Maes, E.G, Wang, L, Taylor, A.B, Hinck, A.P, Lafer, E.M, Sousa, R. | | Deposit date: | 2007-08-10 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis of J cochaperone binding and regulation of Hsp70.
Mol.Cell, 28, 2007
|
|
2QWP
 
 | | Crystal structure of disulfide-bond-crosslinked complex of bovine hsc70 (1-394aa)R171C and bovine Auxilin (810-910aa)D876C in the ADP*Pi form #2 | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Jiang, J, Maes, E.G, Wang, L, Taylor, A.B, Hinck, A.P, Lafer, E.M, Sousa, R. | | Deposit date: | 2007-08-10 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of J cochaperone binding and regulation of Hsp70.
Mol.Cell, 28, 2007
|
|
1JZT
 
 | |
2QWL
 
 | | Crystal structure of bovine hsc70 (1-394aa)in the ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Heat shock cognate 71 kDa protein, ... | | Authors: | Jiang, J, Maes, E.G, Wang, L, Taylor, A.B, Hinck, A.P, Lafer, E.M, Sousa, R. | | Deposit date: | 2007-08-10 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of J cochaperone binding and regulation of Hsp70.
Mol.Cell, 28, 2007
|
|
1YUW
 
 | |
1NZ6
 
 | | Crystal Structure of Auxilin J-Domain | | Descriptor: | Auxilin | | Authors: | Jiang, J, Taylor, A.B, Prasad, K, Ishikawa-Brush, Y, Hart, P.J, Lafer, E.M, Sousa, R. | | Deposit date: | 2003-02-16 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function analysis of the auxilin J-domain reveals an extended Hsc70 interaction interface.
Biochemistry, 42, 2003
|
|
2R7Y
 
 | |
2DLJ
 
 | |
2ESR
 
 | | conserved hypothetical protein- streptococcus pyogenes | | Descriptor: | Methyltransferase, alpha-D-glucopyranose | | Authors: | Jiang, J, Min, T, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-10-26 | | Release date: | 2006-02-07 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of hypothetical protein of Streptococcus Pygenes
To be Published
|
|
6D6V
 
 | | CryoEM structure of Tetrahymena telomerase with telomeric DNA at 4.8 Angstrom resolution | | Descriptor: | DNA (5'-D(P*GP*TP*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*G)-3'), RNA (159-MER), Telomerase associated protein p65, ... | | Authors: | Jiang, J, Wang, Y, Susac, L, Chan, H, Basu, R, Zhou, Z.H, Feigon, J. | | Deposit date: | 2018-04-22 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of Telomerase with Telomeric DNA.
Cell, 173, 2018
|
|
7X3M
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07045 | | Descriptor: | (2~{R})-2-[4-[3,5-bis(chloranyl)phenyl]-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7X3L
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07044 | | Descriptor: | (2~{R})-2-[4-(3-fluoranyl-4-methyl-phenyl)-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
1P32
 
 | | CRYSTAL STRUCTURE OF HUMAN P32, A DOUGHNUT-SHAPED ACIDIC MITOCHONDRIAL MATRIX PROTEIN | | Descriptor: | MITOCHONDRIAL MATRIX PROTEIN, SF2P32 | | Authors: | Jiang, J, Zhang, Y, Krainer, A.R, Xu, R.-M. | | Deposit date: | 1998-11-02 | | Release date: | 1999-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7KGK
 
 | | Crystal structure of synthetic nanobody (Sb16) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | Sb16, Sybody-16, Synthetic Nanobody, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-10-16 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7KLW
 
 | | Crystal structure of synthetic nanobody (Sb45+Sb68) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | SB45, Synthetic Nanobody, SB68, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-11-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
3UM0
 
 | |
3UM3
 
 | |
3UM1
 
 | |
6CT0
 
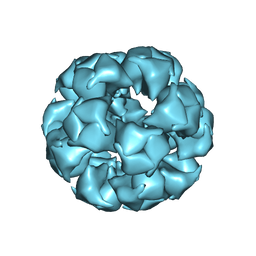 | | Atomic Structure of the E2 Inner Core of Human Pyruvate Dehydrogenase Complex | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | | Authors: | Jiang, J, Baiesc, F.L, Hiromasa, Y, Yu, X, Hui, W.H, Dai, X, Roche, T.E, Zhou, Z.H. | | Deposit date: | 2018-03-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic Structure of the E2 Inner Core of Human Pyruvate Dehydrogenase Complex.
Biochemistry, 57, 2018
|
|
7KGJ
 
 | | Crystal structure of synthetic nanobody (Sb45) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | Sb45, Sybody-45, Synthetic Nanobody, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-10-16 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
3UM2
 
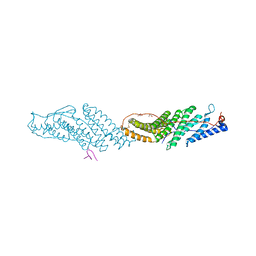 | |
3ULY
 
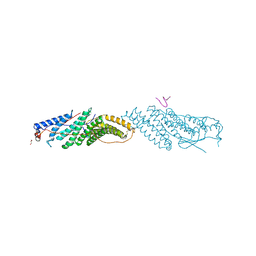 | |
7X3O
 
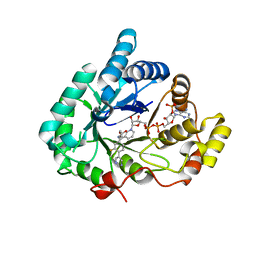 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054 | | Descriptor: | (2~{R})-2-(3-fluoranyl-4-pyrimidin-5-yl-phenyl)butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054
To Be Published
|
|
