5H1Y
 
 | |
5EFX
 
 | | Crystal structure of Rho GTPase regulator | | Descriptor: | Rho guanine nucleotide exchange factor 2 | | Authors: | Jiang, Y, Ouyang, S, Liu, Z.J. | | Deposit date: | 2015-10-26 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Crystal structure of hGEF-H1 PH domain provides insight into incapability in phosphoinositide binding
Biochem.Biophys.Res.Commun., 471, 2016
|
|
5BOB
 
 | | Crystal Structure of the Meningitis Pathogen Streptococcus suis adhesion Fhb | | Descriptor: | GLYCEROL, Translation initiation factor 2 (IF-2 GTPase) | | Authors: | Jiang, Y, Zhang, C, Yu, Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Expression, purification, crystallization and structure determination of the N terminal domain of Fhb, a factor H binding protein from Streptococcus suis.
Biochem.Biophys.Res.Commun., 466, 2015
|
|
3K04
 
 | | Crystal Structure of CNG mimicking NaK mutant, NaK-DTPP, Na+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein NaK, SODIUM ION | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3K0D
 
 | | Crystal Structure of CNG mimicking NaK mutant, NaK-ETPP, K+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein NaK | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3K06
 
 | | Crystal Structure of CNG mimicking NaK mutant, NaK-NTPP, K+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein NaK | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3K0G
 
 | | Crystal Structure of CNG mimicking NaK mutant, NaK-ETPP, Na+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein NaK, SODIUM ION | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3K08
 
 | | Crystal Structure of CNG mimicking NaK mutant, NaK-NTPP, Na+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein NaK, SODIUM ION | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1LNQ
 
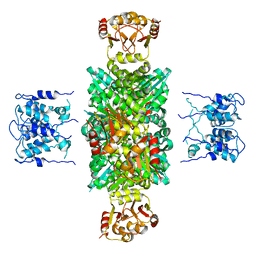 | | CRYSTAL STRUCTURE OF MTHK AT 3.3 A | | Descriptor: | CALCIUM ION, POTASSIUM CHANNEL RELATED PROTEIN | | Authors: | Jiang, Y, Lee, A, Chen, J, Cadene, M, Chait, B.T, Mackinnon, R. | | Deposit date: | 2002-05-03 | | Release date: | 2002-06-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | CRYSTAL STRUCTURE AND MECHANISM OF A CALCIUM-GATED POTASSIUM CHANNEL
Nature, 417, 2002
|
|
6AH8
 
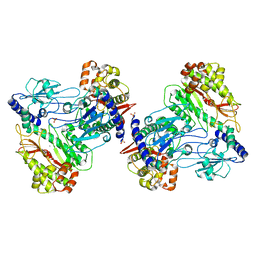 | |
1ORQ
 
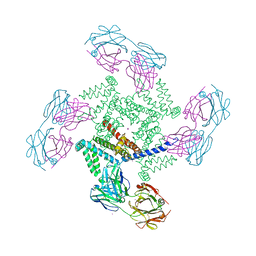 | | X-ray structure of a voltage-dependent potassium channel in complex with an Fab | | Descriptor: | 6E1 Fab heavy chain, 6E1 Fab light chain, CADMIUM ION, ... | | Authors: | Jiang, Y, Lee, A, Chen, J, Ruta, V, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2003-03-14 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a voltage-dependent K+ channel
Nature, 423, 2003
|
|
1ORS
 
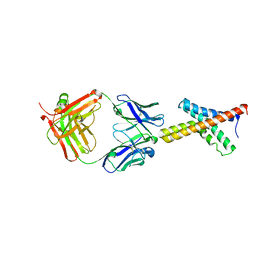 | | X-ray structure of the KvAP potassium channel voltage sensor in complex with an Fab | | Descriptor: | 33H1 Fab heavy chain, 33H1 Fab light chain, potassium channel | | Authors: | Jiang, Y, Lee, A, Chen, J, Ruta, V, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2003-03-14 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of a voltage-dependent K+ channel
Nature, 423, 2003
|
|
2L5F
 
 | | Solution structure of the tandem WW domains from HYPA/FBP11 | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Jiang, Y, Hu, H. | | Deposit date: | 2010-11-01 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Interaction with polyglutamine expanded huntingtin alters cellular distribution and RNA processing of huntingtin yeast two-hybrid protein A (HYPA)
To be Published
|
|
6O6J
 
 | |
6O7A
 
 | |
6PPT
 
 | |
6O7C
 
 | |
6PQ2
 
 | |
6PQM
 
 | |
6PRQ
 
 | |
6PRJ
 
 | |
6PRI
 
 | |
6PQE
 
 | |
6PRP
 
 | |
6PSI
 
 | |
