7BV7
 
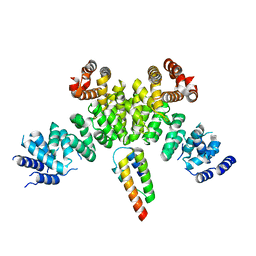 | | INTS3 complexed with INTS6 | | Descriptor: | Integrator complex subunit 3, Integrator complex subunit 6 | | Authors: | Jia, Y, Bharath, S.R, Song, H. | | Deposit date: | 2020-04-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the INTS3/INTS6 complex reveals the functional importance of INTS3 dimerization in DSB repair.
Cell Discov, 7, 2021
|
|
6O7C
 
 | |
6O7A
 
 | |
8TEG
 
 | |
8TEJ
 
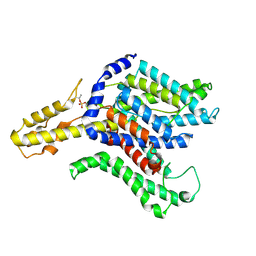 | |
8TEL
 
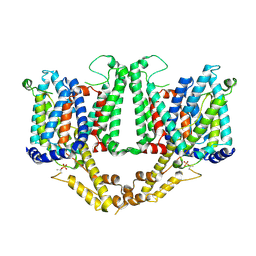 | |
8TEH
 
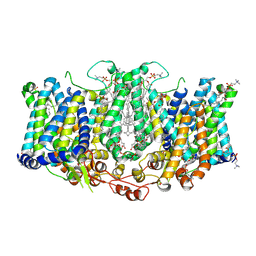 | | Cryo-EM structure of Arabidopsis thaliana Bor1 in lipid nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Boron transporter 1 | | Authors: | Jiang, Y, Jiang, J. | | Deposit date: | 2023-07-06 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structure of borate transporter Bor1 reveals a novel auto-inhibition mechanism for
the SLC4 family
To Be Published
|
|
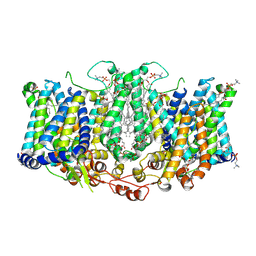
8TEI
 
 | |
8TEM
 
 | |
8TEN
 
 | |
5EFX
 
 | | Crystal structure of Rho GTPase regulator | | Descriptor: | Rho guanine nucleotide exchange factor 2 | | Authors: | Jiang, Y, Ouyang, S, Liu, Z.J. | | Deposit date: | 2015-10-26 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Crystal structure of hGEF-H1 PH domain provides insight into incapability in phosphoinositide binding
Biochem.Biophys.Res.Commun., 471, 2016
|
|
6KND
 
 | |
8JC4
 
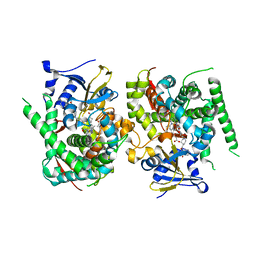 | | Crystal structure of the P450 BM3 heme domain mutant F87A-T268V in complex with Pyd-Pid-Phe and hydroxylamine | | Descriptor: | 1-pyridin-4-ylpiperidine-4-carboxylic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2023-05-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | C(sp3)-H hydroxylation of Broad-Spectrum Alkanes Catalyzed by an Artificial P450 Peroxygenase Driven by omega-Pyridyl Fatty Acyl Amino Acids.
Mol Catal, 550, 2023
|
|
8JC3
 
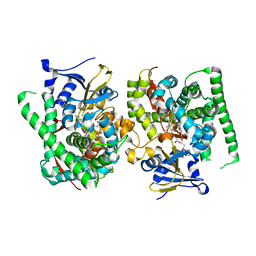 | | Crystal structure of the P450 BM3 heme domain mutant F87A-T268V in complex with Pyd-N-C4-Phe and hydroxylamine | | Descriptor: | 4-(pyridin-4-ylamino)butanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2023-05-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | C(sp3)-H hydroxylation of Broad-Spectrum Alkanes Catalyzed by an Artificial P450 Peroxygenase Driven by omega-Pyridyl Fatty Acyl Amino Acids.
Mol Catal, 550, 2023
|
|
8YTB
 
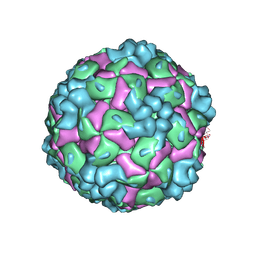 | | Cryo-EM structure of enterovirus A71 empty particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2024-03-25 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM structure of enterovirus A71 empty particle
To Be Published
|
|
8YTJ
 
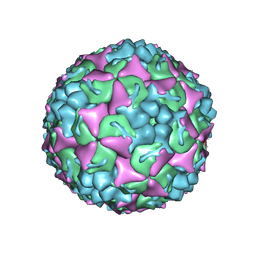 | | Cryo-EM structure of enterovirus A71 mature virion | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2024-03-26 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Cryo-EM structure of enterovirus A71 mature virion in complex with Fab h1A6.2
To Be Published
|
|
4LMM
 
 | | Crystal structure of NHERF1 PDZ1 domain complexed with the CXCR2 C-terminal tail in P21 space group | | Descriptor: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-07-10 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
6PPT
 
 | |
6PQ2
 
 | |
6PQM
 
 | |
6PRQ
 
 | |
6PRJ
 
 | |
6PRP
 
 | |
6PQE
 
 | |
6PRI
 
 | |
