4NJ6
 
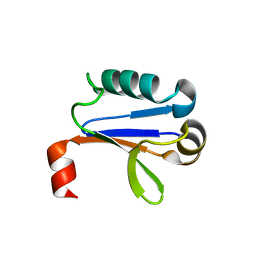 | | PB1 Domain of AtARF7 | | Descriptor: | Auxin response factor 7 | | Authors: | Korasick, D.A, Westfall, C.S, Lee, S.G, Nanao, M, Jez, J.M, Strader, L.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5IUU
 
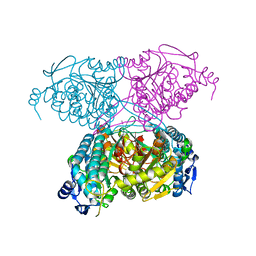 | |
4L39
 
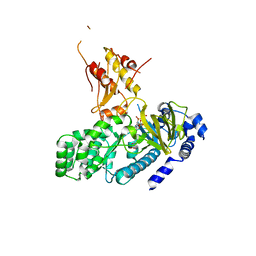 | | Crystal structure of GH3.12 from Arabidopsis thaliana in complex with AMPCPP and salicylate | | Descriptor: | 2-HYDROXYBENZOIC ACID, 4-substituted benzoates-glutamate ligase GH3.12, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Zubieta, C, Jez, J.M, Brown, E, Marcellin, R, Kapp, U, Round, A, Westfall, C. | | Deposit date: | 2013-06-05 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Determination of the GH3.12 protein conformation through HPLC-integrated SAXS measurements combined with X-ray crystallography.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5IUW
 
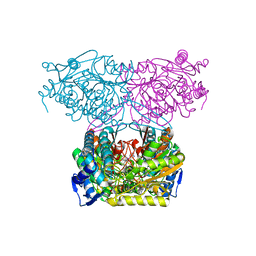 | | Crystal Structure of Indole-3-acetaldehyde Dehydrogenase in complexed with NAD+ and IAA | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Aldehyde dehydrogenase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, S.G, McClerklin, S, Kunkel, B, Jez, J.M. | | Deposit date: | 2016-03-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000.
PLoS Pathog., 14, 2018
|
|
5IUV
 
 | | Crystal Structure of Indole-3-acetaldehyde Dehydrogenase in complexed with NAD+ | | Descriptor: | Aldehyde dehydrogenase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, S.G, McClerklin, S, Kunkel, B, Jez, J.M. | | Deposit date: | 2016-03-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000.
PLoS Pathog., 14, 2018
|
|
7MKU
 
 | |
1CGZ
 
 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH RESVERATROL | | Descriptor: | PROTEIN (CHALCONE SYNTHASE), RESVERATROL | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-26 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1CML
 
 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH MALONYL-COA | | Descriptor: | MALONYL-COENZYME A, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), PROTEIN (CHALCONE SYNTHASE), ... | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-30 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1CHW
 
 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH HEXANOYL-COA | | Descriptor: | HEXANOYL-COENZYME A, PROTEIN (CHALCONE SYNTHASE) | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-30 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1CGK
 
 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH NARINGENIN | | Descriptor: | NARINGENIN, PROTEIN (CHALCONE SYNTHASE) | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-25 | | Release date: | 1999-09-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1BQ6
 
 | | CHALCONE SYNTHASE FROM ALFALFA WITH COENZYME A | | Descriptor: | CHALCONE SYNTHASE, COENZYME A, SULFATE ION | | Authors: | Ferrer, J.-L, Bowman, M.E, Jez, J, Dixon, R, Noel, J.P. | | Deposit date: | 1998-08-21 | | Release date: | 1999-08-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
4PPU
 
 | |
4N69
 
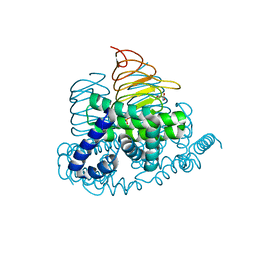 | | Soybean Serine Acetyltransferase Complexed with Serine | | Descriptor: | PHOSPHATE ION, SERINE, Serine Acetyltransferase Apoenzyme | | Authors: | Yi, H, Dey, S, Kumaran, S, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2013-10-11 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of soybean serine acetyltransferase and formation of the cysteine regulatory complex as a molecular chaperone.
J.Biol.Chem., 288, 2013
|
|
4FXP
 
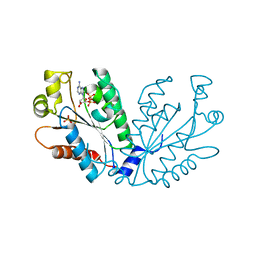 | |
4EPL
 
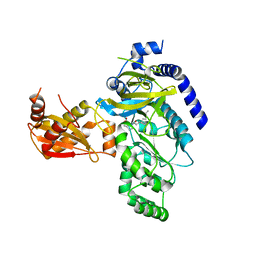 | | Crystal Structure of Arabidopsis thaliana GH3.11 (JAR1) in Complex with JA-Ile | | Descriptor: | Jasmonic acid-amido synthetase JAR1, N-({(1R,2R)-3-oxo-2-[(2Z)-pent-2-en-1-yl]cyclopentyl}acetyl)-L-isoleucine | | Authors: | Westfall, C.S, Zubieta, C, Herrmann, J, Kapp, U, Nanao, M.H, Jez, J.M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
4EPM
 
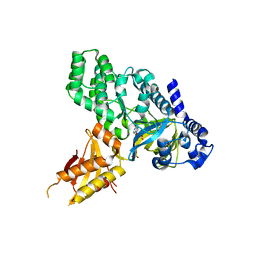 | | Crystal Structure of Arabidopsis GH3.12 (PBS3) in Complex with AMP | | Descriptor: | 4-substituted benzoates-glutamate ligase GH3.12, ADENOSINE MONOPHOSPHATE, SULFATE ION | | Authors: | Westfall, C.S, Zubieta, C, Herrmann, J, Kapp, U, Nanao, M.H, Jez, J.M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
4PPV
 
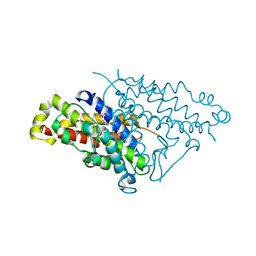 | |
4R6W
 
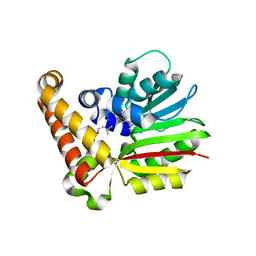 | |
4OMZ
 
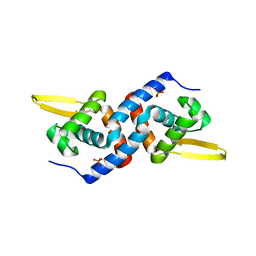 | |
4FGZ
 
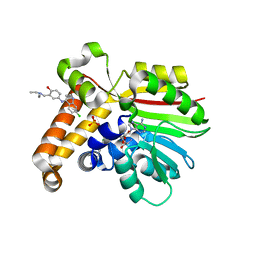 | | Crystal Structure of Phosphoethanolamine Methyltransferase from Plasmodium falciparum in Complex with Amodiaquine | | Descriptor: | 4-[(7-CHLOROQUINOLIN-4-YL)AMINO]-2-[(DIETHYLAMINO)METHYL]PHENOL, PHOSPHATE ION, Phosphoethanolamine N-methyltransferase, ... | | Authors: | Lee, S.G, Alpert, T.D, Jez, J.M. | | Deposit date: | 2012-06-05 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Crystal structure of phosphoethanolamine methyltransferase from Plasmodium falciparum in complex with amodiaquine.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5WMH
 
 | | Arabidopsis thaliana prephenate aminotransferase | | Descriptor: | Bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Holland, C.K, Jez, J.M. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for substrate recognition and inhibition of prephenate aminotransferase from Arabidopsis.
Plant J., 94, 2018
|
|
5W6Y
 
 | | Physcomitrella patens Chorismate Mutase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Chorismate mutase, TRYPTOPHAN | | Authors: | Holland, C.K, Kroll, K, Jez, J.M. | | Deposit date: | 2017-06-18 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Evolution of allosteric regulation in chorismate mutases from early plants.
Biochem. J., 474, 2017
|
|
5WP5
 
 | |
3UIE
 
 | |
5WP4
 
 | |
