5B26
 
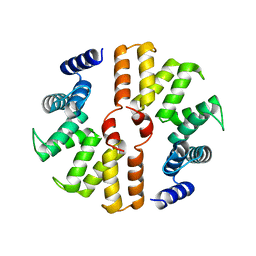 | | Crystal structure of mouse SEL1L | | Descriptor: | Protein sel-1 homolog 1 | | Authors: | Jeong, H, Lee, C. | | Deposit date: | 2016-01-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of SEL1L: Insight into the roles of SLR motifs in ERAD pathway
Sci Rep, 6, 2016
|
|
7USY
 
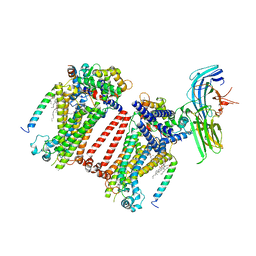 | | Structure of C. elegans TMC-1 complex with ARRD-6 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jeong, H, Clark, S, Gouaux, E. | | Deposit date: | 2022-04-26 | | Release date: | 2022-10-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structures of the TMC-1 complex illuminate mechanosensory transduction.
Nature, 610, 2022
|
|
7USX
 
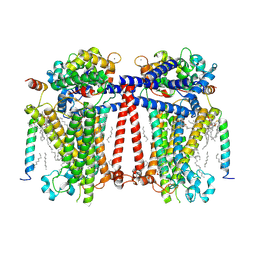 | | Structure of Contracted C. elegans TMC-1 complex | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jeong, H, Clark, S, Gouaux, E. | | Deposit date: | 2022-04-26 | | Release date: | 2022-10-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structures of the TMC-1 complex illuminate mechanosensory transduction.
Nature, 610, 2022
|
|
7USW
 
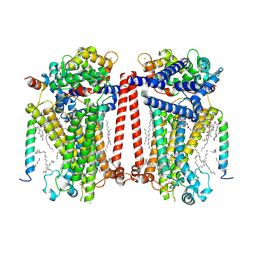 | | Structure of Expanded C. elegans TMC-1 complex | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jeong, H, Clark, S, Gouaux, E. | | Deposit date: | 2022-04-26 | | Release date: | 2022-10-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the TMC-1 complex illuminate mechanosensory transduction.
Nature, 610, 2022
|
|
5GYD
 
 | | Crystal Structure of Mdm12 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Mitochondrial distribution and morphology protein 12 | | Authors: | Jeong, H, Park, J, Lee, C. | | Deposit date: | 2016-09-22 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Crystal structure of Mdm12 reveals the architecture and dynamic organization of the ERMES complex
EMBO Rep., 17, 2016
|
|
5GYK
 
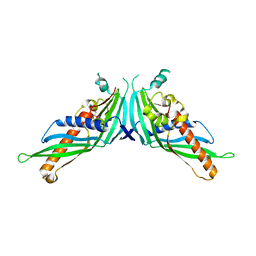 | | Crystal Structure of Mdm12-deletion mutant | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Mitochondrial distribution and morphology protein 12 | | Authors: | Jeong, H, Park, J, Lee, C. | | Deposit date: | 2016-09-22 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.596 Å) | | Cite: | Crystal structure of Mdm12 reveals the architecture and dynamic organization of the ERMES complex
EMBO Rep., 17, 2016
|
|
7FDJ
 
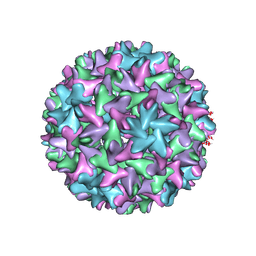 | | Engineered Hepatitis B virus core antigen with short linker T=4 | | Descriptor: | Capsid protein,Immunoglobulin G-binding protein A | | Authors: | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | Deposit date: | 2021-07-16 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
7EP6
 
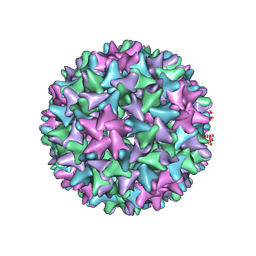 | | Engineered Hepatitis B virus core antigen T=4 | | Descriptor: | Capsid protein,Immunoglobulin G-binding protein A | | Authors: | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | Deposit date: | 2021-04-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
7EOY
 
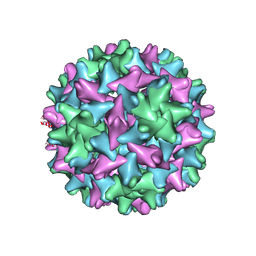 | | Engineered Hepatitis B virus core antigen T=3 | | Descriptor: | Capsid protein,Immunoglobulin G-binding protein A | | Authors: | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | Deposit date: | 2021-04-24 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
5XJG
 
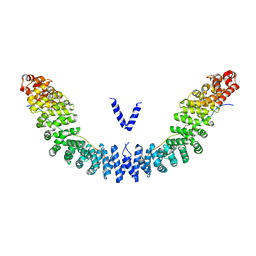 | | Crystal structure of Vac8p bound to Nvj1p | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Nucleus-vacuole junction protein 1, ... | | Authors: | Jeong, H, Park, J, Jun, Y, Lee, C. | | Deposit date: | 2017-05-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insight into the nucleus-vacuole junction based on the Vac8p-Nvj1p crystal structure.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5Y3N
 
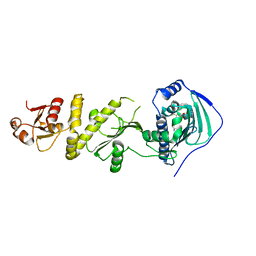 | | Structure of TRAP1 complexed with DN401 | | Descriptor: | 1-[(6-bromanyl-1,3-benzodioxol-5-yl)methyl]-4-chloranyl-pyrazolo[3,4-d]pyrimidin-6-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Jeong, H, Park, H.K, Kang, S, Kang, B.H, Lee, C. | | Deposit date: | 2017-07-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Paralog Specificity Determines Subcellular Distribution, Action Mechanism, and Anticancer Activity of TRAP1 Inhibitors.
J. Med. Chem., 60, 2017
|
|
5Y3O
 
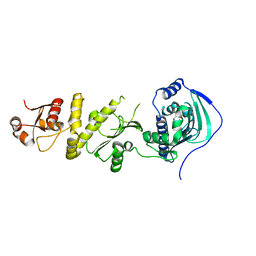 | | Structure of TRAP1 complexed with DN320 | | Descriptor: | 4-chloranyl-1-[(4-methoxy-3,5-dimethyl-pyridin-2-yl)methyl]pyrazolo[3,4-d]pyrimidin-6-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Jeong, H, Park, H.K, Kang, S, Kang, B.H, Lee, C. | | Deposit date: | 2017-07-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Paralog Specificity Determines Subcellular Distribution, Action Mechanism, and Anticancer Activity of TRAP1 Inhibitors.
J. Med. Chem., 60, 2017
|
|
5YK6
 
 | | Crystal Structure of Mmm1 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Maintenance of mitochondrial morphology protein 1 | | Authors: | Jeong, H, Park, J, Lee, C. | | Deposit date: | 2017-10-12 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Mmm1 and Mdm12-Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5YK7
 
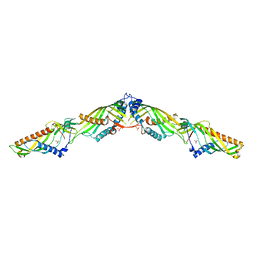 | | Crystal Structure of Mdm12-Mmm1 complex | | Descriptor: | Maintenance of mitochondrial morphology protein 1, Mitochondrial distribution and morphology protein 12, PHOSPHATE ION | | Authors: | Jeong, H, Park, J, Lee, C. | | Deposit date: | 2017-10-12 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.799 Å) | | Cite: | Crystal structures of Mmm1 and Mdm12-Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7WM5
 
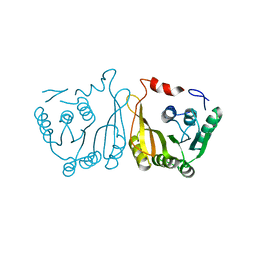 | |
7WM6
 
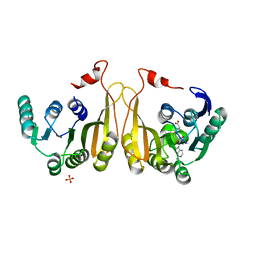 | |
7XM8
 
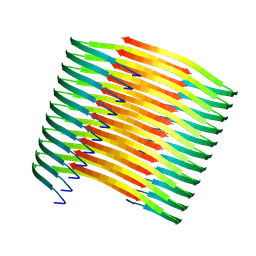 | | Glucagon amyloid fibril | | Descriptor: | Glucagon | | Authors: | Jeong, H, Lin, Y, Lee, Y.-H. | | Deposit date: | 2022-04-25 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomistic zipper-like amyloid structure of full-length glucagon
To Be Published
|
|
7EPP
 
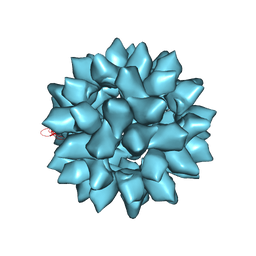 | |
8TKP
 
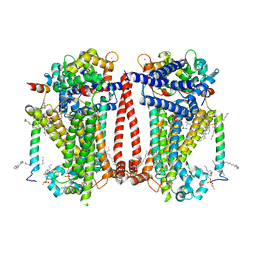 | | Structure of the C. elegans TMC-2 complex | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clark, S, Jeong, H, Goehring, A, Posert, R, Gouaux, E. | | Deposit date: | 2023-07-25 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The structure of the Caenorhabditis elegans TMC-2 complex suggests roles of lipid-mediated subunit contacts in mechanosensory transduction.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8GVW
 
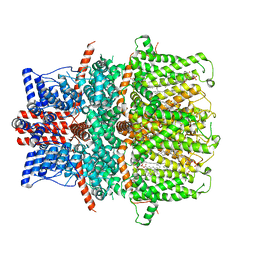 | | Cryo-EM structure of the human TRPC5 ion channel in lipid nanodiscs, class2 | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2022-09-16 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Molecular architecture of the G alpha i -bound TRPC5 ion channel.
Nat Commun, 14, 2023
|
|
8GVX
 
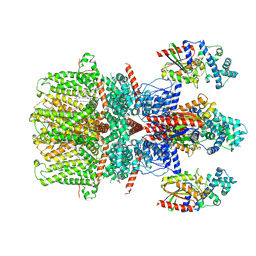 | | Cryo-EM structure of the human TRPC5 ion channel in complex with G alpha i3 subunits, class2 | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2022-09-16 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Molecular architecture of the G alpha i -bound TRPC5 ion channel.
Nat Commun, 14, 2023
|
|
8WPL
 
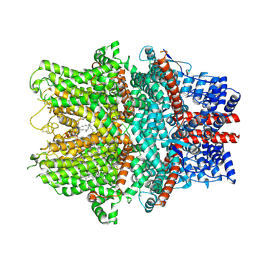 | | Cryo-EM structure of the human TRPC1/C4 heteromer | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Cryo-EM structure of the heteromeric TRPC1/TRPC4 channel.
Nat.Struct.Mol.Biol., 2024
|
|
8WPN
 
 | | Cryo-EM structure of the human TRPC4 in lipid nanodiscs | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Cryo-EM structure of the heteromeric TRPC1/TRPC4 channel.
Nat.Struct.Mol.Biol., 2024
|
|
8WPM
 
 | | Cryo-EM structure of the human TRPC1/C4 heteromer in complex with Pico145 | | Descriptor: | 7-[(4-chlorophenyl)methyl]-3-methyl-1-(3-oxidanylpropyl)-8-[3-(trifluoromethyloxy)phenoxy]purine-2,6-dione, CALCIUM ION, Short transient receptor potential channel 1, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Cryo-EM structure of the heteromeric TRPC1/TRPC4 channel.
Nat.Struct.Mol.Biol., 2024
|
|
8HKP
 
 | | Structurally hetero-junctional human Cx36/GJD2 gap junction channel in detergents (C6 symmetry) | | Descriptor: | Gap junction delta-2 protein, Lauryl Maltose Neopentyl Glycol | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-11-27 | | Release date: | 2023-03-22 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
