5VXF
 
 | |
5VXH
 
 | |
6B2U
 
 | | Crystal structure of Xanthomonas campestris OleA H285N with Cerulenin | | 分子名称: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[ACP] synthase III, ... | | 著者 | Jensen, M.R, Goblirsch, B.R, Esler, M.A, Christenson, J.K, Mohamed, F.A, Wackett, L.P, Wilmot, C.M. | | 登録日 | 2017-09-20 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | The role of OleA His285 in substrate coordination of long-chain acyl-CoA
To be published
|
|
6B2R
 
 | | Crystal structure of Xanthomonas campestris OleA H285A | | 分子名称: | 3-oxoacyl-[ACP] synthase III, GLYCEROL | | 著者 | Jensen, M.R, Goblirsch, B.R, Esler, M.A, Christenson, J.K, Mohamed, F.A, Wackett, L.P, Wilmot, C.M. | | 登録日 | 2017-09-20 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | The role of OleA His285 in orchestration of long-chain acyl-coenzyme A substrates.
FEBS Lett., 592, 2018
|
|
6B2S
 
 | | Crystal structure of Xanthomonas campestris OleA H285N | | 分子名称: | 3-oxoacyl-[ACP] synthase III, GLYCEROL, PHOSPHATE ION | | 著者 | Jensen, M.R, Goblirsch, B.R, Esler, M.A, Christenson, J.K, Mohamed, F.A, Wackett, L.P, Wilmot, C.M. | | 登録日 | 2017-09-20 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The role of OleA His285 in orchestration of long-chain acyl-coenzyme A substrates.
FEBS Lett., 592, 2018
|
|
6B2T
 
 | | Crystal structure of Xanthomonas campestris OleA H285D | | 分子名称: | 3-oxoacyl-[ACP] synthase III, GLYCEROL, PHOSPHATE ION | | 著者 | Jensen, M.R, Goblirsch, B.R, Esler, M.A, Christenson, J.K, Mohamed, F.A, Wackett, L.P, Wilmot, C.M. | | 登録日 | 2017-09-20 | | 公開日 | 2018-02-28 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The role of OleA His285 in orchestration of long-chain acyl-coenzyme A substrates.
FEBS Lett., 592, 2018
|
|
5VXE
 
 | | Crystal structure of Xanthomonas campestris OleA E117A bound with Cerulenin | | 分子名称: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[ACP] synthase III, ... | | 著者 | Jensen, M.R, Wilmot, C.M. | | 登録日 | 2017-05-23 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | OleA Glu117 is key to condensation of two fatty-acyl coenzyme A substrates in long-chain olefin biosynthesis.
Biochem. J., 474, 2017
|
|
5VXG
 
 | | Crystal structure of Xanthomonas campestris OleA E117Q bound with Cerulenin | | 分子名称: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[ACP] synthase III, ... | | 著者 | Jensen, M.R, Wilmot, C.M. | | 登録日 | 2017-05-23 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | OleA Glu117 is key to condensation of two fatty-acyl coenzyme A substrates in long-chain olefin biosynthesis.
Biochem. J., 474, 2017
|
|
5VXD
 
 | |
5VXI
 
 | | Crystal structure of Xanthomonas campestris OleA E117D bound with Cerulenin | | 分子名称: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[ACP] synthase III, ... | | 著者 | Jensen, M.R, Wilmot, C.M. | | 登録日 | 2017-05-23 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | OleA Glu117 is key to condensation of two fatty-acyl coenzyme A substrates in long-chain olefin biosynthesis.
Biochem. J., 474, 2017
|
|
3SWS
 
 | |
3RMZ
 
 | |
3RN0
 
 | |
3SJL
 
 | |
3ORV
 
 | |
3SVW
 
 | |
3L4O
 
 | |
3L4M
 
 | |
3RN1
 
 | |
3SXT
 
 | |
6TRI
 
 | | CI-MOR repressor-antirepressor complex of the temperate bacteriophage TP901-1 from Lactococcus lactis | | 分子名称: | CI, MOR, SULFATE ION | | 著者 | Rasmussen, K.K, Blackledge, M, Herrmann, T, Jensen, M.R, Lo Leggio, L. | | 登録日 | 2019-12-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.277 Å) | | 主引用文献 | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TO6
 
 | | Solution structure of the modulator of repression (MOR) of the temperate bacteriophage TP901-1 from Lactococcus lactis | | 分子名称: | MOR | | 著者 | Rasmussen, K.K, Blackledge, M, Herrmann, T, Lo Leggio, L, Jensen, M.R. | | 登録日 | 2019-12-11 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8RPP
 
 | |
4XW2
 
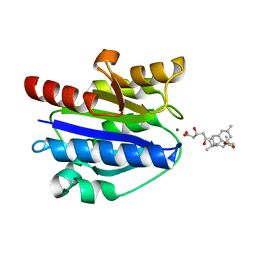 | | Structural basis for simvastatin competitive antagonism of complement receptor 3 | | 分子名称: | Integrin alpha-M, MAGNESIUM ION, Simvastatin acid | | 著者 | Bajic, G, Jensen, M.R, Vorup-Jensen, T, Andersen, G.R. | | 登録日 | 2015-01-28 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural Basis for Simvastatin Competitive Antagonism of Complement Receptor 3.
J.Biol.Chem., 291, 2016
|
|
4FCR
 
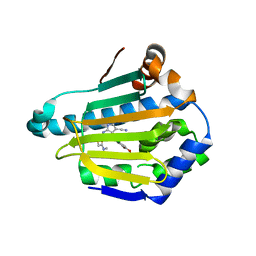 | | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization | | 分子名称: | 2-{[4-(2-chloro-4,5-dimethoxyphenyl)-5-cyano-7H-pyrrolo[2,3-d]pyrimidin-2-yl]sulfanyl}-N,N-dimethylacetamide, Heat shock protein HSP 90-alpha | | 著者 | Davies, N.G, Browne, H, Davis, B, Foloppe, N, Geoffrey, S, Gibbons, B, Hart, T, Drysdale, M.J, Mansell, H, Massey, A, Matassova, N, Moore, J.D, Murray, J, Pratt, R, Ray, S, Roughley, S.D, Jensen, M.R, Schoepfer, J, Scriven, K, Simmonite, H, Stokes, S, Surgenor, A, Webb, P, Wright, L, Brough, P. | | 登録日 | 2012-05-25 | | 公開日 | 2012-10-24 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.698 Å) | | 主引用文献 | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization.
Bioorg.Med.Chem., 20, 2012
|
|
