1QJV
 
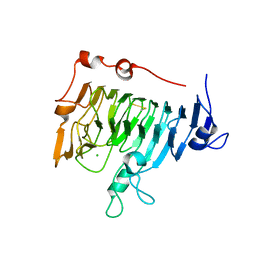 | | Pectin methylesterase PemA from Erwinia chrysanthemi | | Descriptor: | CHLORIDE ION, PECTIN METHYLESTERASE | | Authors: | Jenkins, J, Mayans, O, Smith, D, Worboys, K, Pickersgill, R. | | Deposit date: | 1999-07-05 | | Release date: | 2000-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Three-Dimensional Structure of Erwinia Chrysanthemi Pectin Methylesterase Reveals a Novel Esterase Active Site
J.Mol.Biol., 305, 2001
|
|
7REX
 
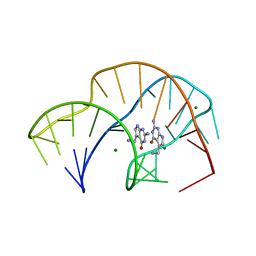 | |
3Q50
 
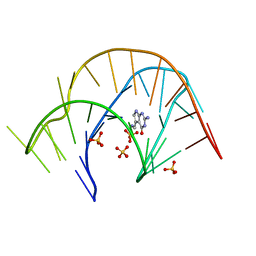 | |
7LHX
 
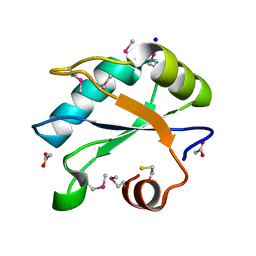 | | Human U1A protein with F37M and F77M mutations for improved phasing | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, SODIUM ION, ... | | Authors: | Jenkins, J.L, Lippa, G.M, Wedekind, J.E. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Affinity and Structural Analysis of the U1A RNA Recognition Motif with Engineered Methionines to Improve Experimental Phasing
Crystals, 11, 2021
|
|
1U8F
 
 | |
4TU9
 
 | | STRUCTURE OF U2AF65 VARIANT WITH BRU5G6 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*DG*U)-3'), GLYCEROL, ... | | Authors: | Jenkins, J.L, McLaughlin, K.J, Agrawal, A.A, Kielkopf, C.L. | | Deposit date: | 2014-06-24 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1RU4
 
 | | Crystal structure of pectate lyase Pel9A | | Descriptor: | CALCIUM ION, Pectate lyase | | Authors: | Jenkins, J, Shevchik, V.E, Hugouvieux-Cotte-Pattat, N, Pickersgill, R.W. | | Deposit date: | 2003-12-11 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of pectate lyase Pel9A from Erwinia chrysanthemi
J.Biol.Chem., 279, 2004
|
|
6VUH
 
 | | APO PreQ1 riboswitch aptamer grown in Mn2+ | | Descriptor: | MANGANESE (II) ION, PREQ1 RIBOSWITCH | | Authors: | Jenkins, J.L, Wedekind, J.E. | | Deposit date: | 2020-02-15 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Analysis of a preQ1-I riboswitch in effector-free and bound states reveals a metabolite-programmed nucleobase-stacking spine that controls gene regulation.
Nucleic Acids Res., 48, 2020
|
|
6VUI
 
 | | Metabolite-bound PreQ1 riboswitch with Mn2+ | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, MANGANESE (II) ION, PREQ1 RIBOSWITCH | | Authors: | Jenkins, J.L, Wedekind, J.E. | | Deposit date: | 2020-02-15 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.681 Å) | | Cite: | Analysis of a preQ1-I riboswitch in effector-free and bound states reveals a metabolite-programmed nucleobase-stacking spine that controls gene regulation.
Nucleic Acids Res., 48, 2020
|
|
2GPE
 
 | |
2G82
 
 | |
6NSX
 
 | |
3VAK
 
 | | Structure of U2AF65 variant with BrU5 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*UP*U)-3'), GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAF
 
 | | Structure of U2AF65 variant with BrU3 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(*UP*UP*(BRU)P*(BRU)P*UP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAM
 
 | | Structure of U2AF65 variant with BrU5C2 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*CP*UP*UP*(BRU)P*UP*U)-3'), GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAJ
 
 | | Structure of U2AF65 variant with BrU5C6 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*CP*U)-3'), GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAI
 
 | | Structure of U2AF65 variant with BrU3C5 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(*UP*UP*(BRU)P*UP*CP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Frato, K.H, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAG
 
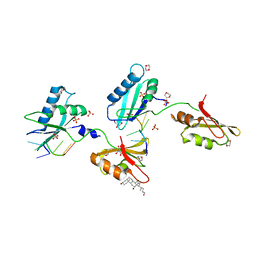 | | Structure of U2AF65 variant with BrU3C2 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(*U*CP*(BRU)P*UP*UP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAH
 
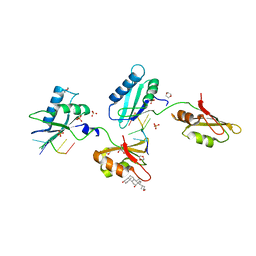 | | Structure of U2AF65 variant with BrU3C4 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(P*UP*UP*(BRU)P*CP*UP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAL
 
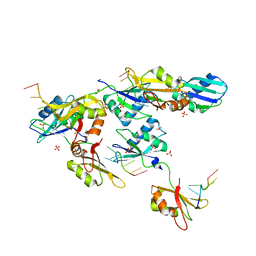 | | Structure of U2AF65 variant with BrU5C1 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*C*UP*UP*UP*(BRU)P*UP*U)-3'), SULFATE ION, ... | | Authors: | Jenkins, J.L, Frato, K.H, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
7S3B
 
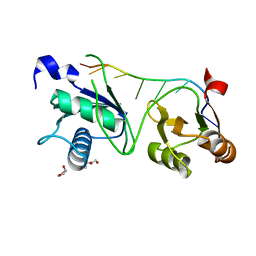 | |
7S3A
 
 | |
7SLI
 
 | | Engineered sperm whale myoglobin-based carbene transferase MbBTIC-C2 | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Jenkins, J.L, Vargas, D, Ren, X, Fasan, R. | | Deposit date: | 2021-10-24 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biocatalytic strategy for the construction of sp 3 -rich polycyclic compounds from directed evolution and computational modelling.
Nat.Chem., 16, 2024
|
|
7SLH
 
 | | Engineered sperm whale myoglobin-based carbene transferase MbBTIC-C3 | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Jenkins, J.L, Vargas, D, Ren, X, Fasan, R. | | Deposit date: | 2021-10-24 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Biocatalytic strategy for the construction of sp 3 -rich polycyclic compounds from directed evolution and computational modelling.
Nat.Chem., 16, 2024
|
|
5SYT
 
 | | Crystal Structure of ZMPSTE24 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CAAX prenyl protease 1 homolog, ... | | Authors: | Clark, K, Jenkins, J.L, Fedoriw, N, Dumont, M.E, Membrane Protein Structural Biology Consortium (MPSBC) | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human CaaX protease ZMPSTE24 expressed in yeast: Structure and inhibition by HIV protease inhibitors.
Protein Sci., 26, 2017
|
|
