6MGY
 
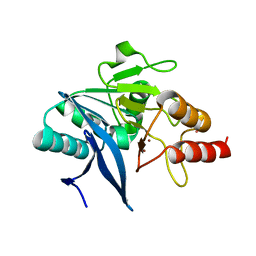 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 5 from Klebsiella pneumoniae | | Descriptor: | GLYCEROL, Metallo-beta-lactamase NDM-5, POTASSIUM ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 5 from Klebsiella pneumoniae
To Be Published
|
|
5KCK
 
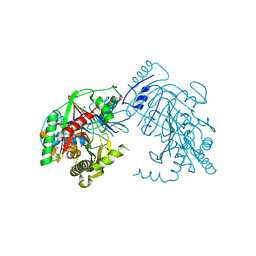 | | Crystal structure of anthranilate synthase component I from Streptococcus pneumoniae TIGR4 | | Descriptor: | Anthranilate synthase component I, GLYCEROL | | Authors: | Chang, C, Michalska, K, Bigelow, L, Jedrzejczak, R, ANDERSON, W.F, JOACHIMIAK, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-06 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of anthranilate synthase component I from Streptococcus pneumoniae TIGR4
To Be Published
|
|
6MH0
 
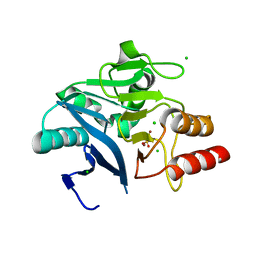 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 3 from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, Metallo-beta-lactamase NDM-3, SULFATE ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 3 from Klebsiella pneumoniae
To Be Published
|
|
3SOZ
 
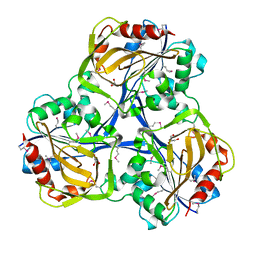 | | Cytoplasmic Protein STM1381 from Salmonella typhimurium LT2 | | Descriptor: | Cytoplasmic Protein STM1381, GLYCEROL | | Authors: | Joachimiak, A, Duke, N.E.C, Jedrzejczak, R, Li, H, Adkins, J, Brown, R, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2011-06-30 | | Release date: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cytoplasmic Protein STM1381 from Salmonella typhimurium LT2
To be Published
|
|
6NJC
 
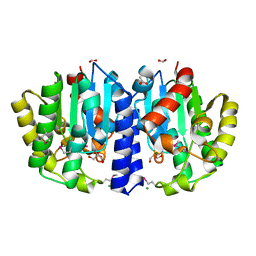 | | Crystal Structure of the Sialate O-acetylesterase from Bacteroides vulgatus | | Descriptor: | ACETIC ACID, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Li, H, Biglow, L, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-01-03 | | Release date: | 2019-01-16 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Sialate O-acetylesterase from Bacteroides vulgatus
To Be Published
|
|
6NKF
 
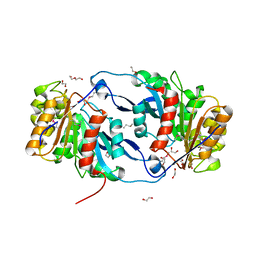 | | Crystal Structure of the Lipase Lip_vut4 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, 2-BUTANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.232 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut4 from Goat Rumen metagenome.
To Be Published
|
|
6WIQ
 
 | | Crystal structure of the co-factor complex of NSP7 and the C-terminal domain of NSP8 from SARS CoV-2 | | Descriptor: | Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
6WQD
 
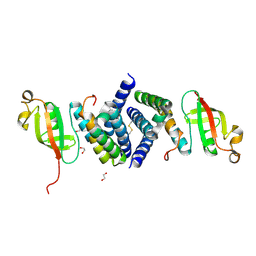 | | The 1.95 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | Authors: | Kim, Y, Wilamowski, M, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
6MGZ
 
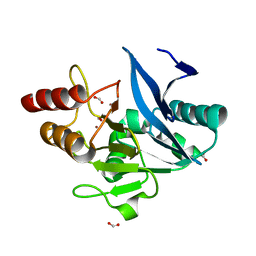 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 4 from Klebsiella pneumoniae | | Descriptor: | FORMIC ACID, MAGNESIUM ION, NDM-4, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 4 from Klebsiella pneumoniae
To Be Published
|
|
6XOA
 
 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with C145S mutation | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with C145S mutation
To Be Published
|
|
6MGX
 
 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 6 Klebsiella pneumoniae | | Descriptor: | Metallo-beta-lactamase, SULFATE ION, ZINC ION | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 6 Klebsiella pneumoniae
To Be Published
|
|
3SQN
 
 | | Putative Mga family transcriptional regulator from Enterococcus faecalis | | Descriptor: | Conserved domain protein | | Authors: | Osipiuk, J, Wu, R, Jedrzejczak, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-07-05 | | Release date: | 2011-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Putative Mga family transcriptional regulator from Enterococcus faecalis.
To be Published
|
|
6NKC
 
 | | Crystal Structure of the Lipase Lip_vut1 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut1 from Goat Rumen metagenome.
To Be Published
|
|
6NKG
 
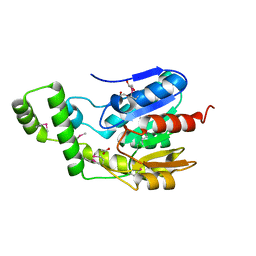 | | Crystal Structure of the Lipase Lip_vut5 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Lip_vut5, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut5 from Goat Rumen metagenome.
To Be Published
|
|
6NKD
 
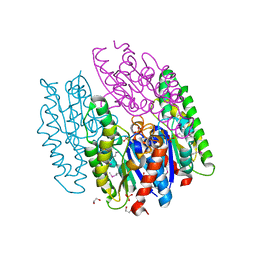 | | Crystal Structure of the Lipase Lip_vut3 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut3 from Goat Rumen metagenome.
To Be Published
|
|
3T9Y
 
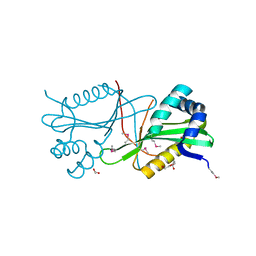 | | Crystal structure of GNAT family acetyltransferase Staphylococcus aureus subsp. aureus USA300_TCH1516 | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase, GNAT family, ... | | Authors: | Chang, C, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-03 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GNAT family acetyltransferase Staphylococcus aureus subsp. aureus USA300_TCH1516
To be Published
|
|
7KB3
 
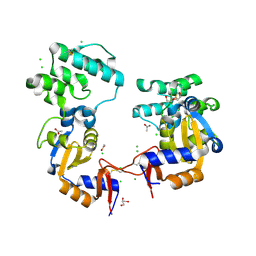 | | The structure of a sensor domain of a histidine kinase (VxrA) from Vibrio cholerae O1 biovar eltor str. N16961, 2nd form | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 2021
|
|
6XKF
 
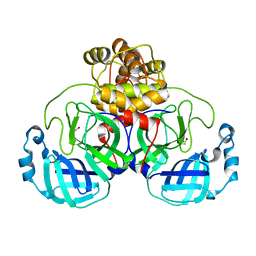 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with oxidized Cys145 (Sulfenic acid cysteine). | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Coates, L, Kovalevskyi, A.Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with oxidized Cys145 (Sulfenic acid cysteine).
To Be Published
|
|
6XIP
 
 | | The 1.5 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
6XKH
 
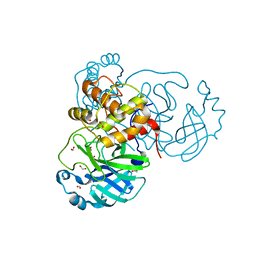 | | THE 1.28A CRYSTAL STRUCTURE OF 3CL MAINPRO OF SARS-COV-2 WITH OXIDIZED C145 (sulfinic acid cysteine) | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, ACETATE ION, ... | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Coates, L, Kovalevsky, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | THE 1.28A CRYSTAL STRUCTURE OF 3CL MAINPRO OF SARS-COV-2 WITH OXIDIZED C145 (sulfinic acid cysteine)
To Be Published
|
|
3CAN
 
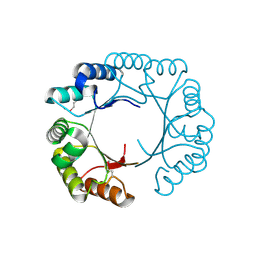 | | Crystal structure of a domain of pyruvate-formate lyase-activating enzyme from Bacteroides vulgatus ATCC 8482 | | Descriptor: | Pyruvate-formate lyase-activating enzyme | | Authors: | Nocek, B, Hendricks, R, Hatzos, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a domain of pyruvate-formate lyase-activating enzyme from Bacteroides vulgatus ATCC 8482.
To be Published
|
|
7KB7
 
 | | THE STRUCTURE OF A SENSOR DOMAIN OF A HISTIDINE KINASE (VxrA) FROM VIBRIO CHOLERAE O1 BIOVAR ELTOR STR. N16961, N239-T240 deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Membrane Proteins of Infectious Diseases (MPID) | | Deposit date: | 2020-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 2021
|
|
7KB9
 
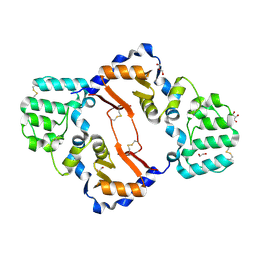 | | THE STRUCTURE OF A SENSOR DOMAIN OF A HISTIDINE KINASE (VxrA) FROM VIBRIO CHOLERAE O1 BIOVAR ELTOR STR. N16961, D238-T240 deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Sensor histidine kinase | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 2021
|
|
3D0K
 
 | | Crystal structure of the LpqC, poly(3-hydroxybutyrate) depolymerase from Bordetella parapertussis | | Descriptor: | CHLORIDE ION, FORMIC ACID, Putative poly(3-hydroxybutyrate) depolymerase LpqC, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-01 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of the LpqC, Poly(3-hydroxybutyrate) Depolymerase from Bordetella parapertussis.
To be Published
|
|
3D3S
 
 | | Crystal structure of L-2,4-diaminobutyric acid acetyltransferase from Bordetella parapertussis | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, GLYCEROL, L-2,4-diaminobutyric acid acetyltransferase, ... | | Authors: | Kim, Y, Volkart, L, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of L-2,4-diaminobutyric acid acetyltransferase from Bordetella parapertussis.
To be Published
|
|
