4HKY
 
 | | New Delhi Metallo-beta-Lactamase-1, Complexed with Cd and Faropenem | | 分子名称: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, (5R,6S)-6-(1-hydroxyethyl)-7-oxo-3-[(2R)-oxolan-2-yl]-4-thia-1-azabicyclo[3.2.0]hept-2-ene-2-carboxylic acid, Beta-lactamase NDM-1, ... | | 著者 | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2012-10-15 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.004 Å) | | 主引用文献 | New Delhi Metallo-beta-Lactamase-1, Complexed with Cd and Faropenem
To be Published
|
|
4GMD
 
 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with AZT Monophosphate | | 分子名称: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2012-08-15 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with AZT Monophosphate
To be Published
|
|
4EDH
 
 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with ADP,TMP and Mg. | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2012-03-27 | | 公開日 | 2012-05-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with ADP,TMP and Mg.
To be Published
|
|
4ESH
 
 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with deoxythymidine. | | 分子名称: | THYMIDINE, Thymidylate kinase | | 著者 | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2012-04-23 | | 公開日 | 2012-05-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with deoxythymidine.
To be Published
|
|
4ER9
 
 | | Crystal structure of cytochrome b562 from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | 著者 | Tan, K, Hatzos-Skintges, C, Jedrzejczak, R, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | 登録日 | 2012-04-19 | | 公開日 | 2012-05-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.897 Å) | | 主引用文献 | Crystal structure of cytochrome b562 from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S
To be Published
|
|
4E5U
 
 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with thymidine monophosphate. | | 分子名称: | CALCIUM ION, THYMIDINE-5'-PHOSPHATE, Thymidylate kinase | | 著者 | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2012-03-14 | | 公開日 | 2012-03-28 | | 実験手法 | X-RAY DIFFRACTION (1.812 Å) | | 主引用文献 | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with thymidine monophosphate.
To be Published
|
|
4GYQ
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 D223A mutant from Klebsiella pneumoniae | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase NDM-1, MAGNESIUM ION | | 著者 | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2012-09-05 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.351 Å) | | 主引用文献 | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 D223A mutant from Klebsiella pneumoniae
To be Published, 2012
|
|
4HAO
 
 | | Crystal Structure of Inorganic Polyphosphate/ATP-NAD Kinase from Yersinia pestis CO92 | | 分子名称: | ACETIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase, SULFATE ION | | 著者 | Kim, Y, Maltseva, N, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-09-27 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.551 Å) | | 主引用文献 | Crystal Structure of Inorganic Polyphosphate/ATP-NAD Kinase from Yersinia pestis CO92
To be Published
|
|
4HCJ
 
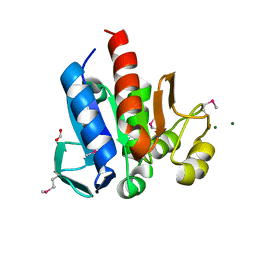 | | Crystal Structure of ThiJ/PfpI Domain Protein from Brachyspira murdochii | | 分子名称: | CHLORIDE ION, FORMIC ACID, MAGNESIUM ION, ... | | 著者 | Kim, Y, Bigelow, L, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-09-30 | | 公開日 | 2012-10-24 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Crystal Structure of ThiJ/PfpI Domain Protein from Brachyspira murdochii
To be Published
|
|
4GYU
 
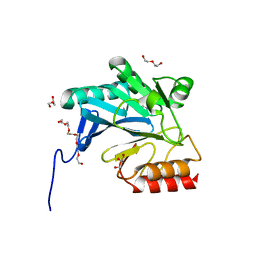 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 A121F mutant from Klebsiella pneumoniae | | 分子名称: | Beta-lactamase NDM-1, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2012-09-05 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 A121F mutant from Klebsiella pneumoniae
To be Published, 2012
|
|
5T87
 
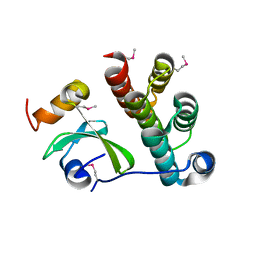 | | Crystal structure of CDI complex from Cupriavidus taiwanensis LMG 19424 | | 分子名称: | CdiA toxin, CdiI immunity protein | | 著者 | Michalska, K, Joachimiak, G, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2016-09-06 | | 公開日 | 2017-09-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Target highlights from the first post-PSI CASP experiment (CASP12, May-August 2016).
Proteins, 86 Suppl 1, 2018
|
|
4PF1
 
 | | Crystal structure of aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon | | 分子名称: | GLYCEROL, Peptidase S15/CocE/NonD, TRIETHYLENE GLYCOL | | 著者 | Michalska, K, Chhor, G, Fayman, K, Endres, M, Jedrzejczak, R, Babnigg, G, Steen, A, Lloyd, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-04-25 | | 公開日 | 2014-06-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | New aminopeptidase from "microbial dark matter" archaeon.
FASEB J., 29, 2015
|
|
4H3U
 
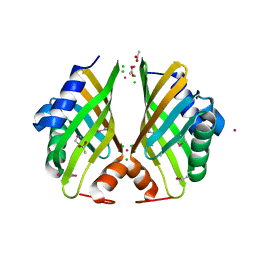 | | Crystal structure of hypothetical protein with ketosteroid isomerase-like protein fold from Catenulispora acidiphila DSM 44928 | | 分子名称: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-09-14 | | 公開日 | 2012-10-03 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Structural characterization of a hypothetical protein: a potential agent involved in trimethylamine metabolism in Catenulispora acidiphila.
J.Struct.Funct.Genom., 15, 2014
|
|
4HVN
 
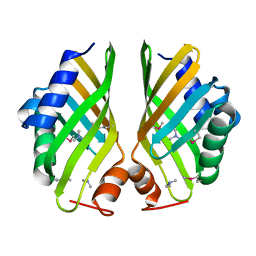 | | Crystal structure of hypothetical protein with ketosteroid isomerase-like protein fold from Catenulispora acidiphila DSM 44928 in complex with Trimethylamine. | | 分子名称: | N,N-dimethylmethanamine, hypothetical protein | | 著者 | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-11-06 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural characterization of a hypothetical protein: a potential agent involved in trimethylamine metabolism in Catenulispora acidiphila.
J.Struct.Funct.Genom., 15, 2014
|
|
4GPN
 
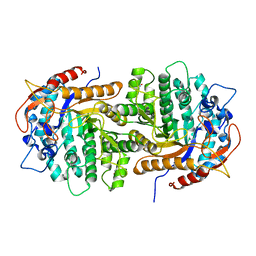 | | The crystal structure of 6-P-beta-D-Glucosidase (E375Q mutant) from Streptococcus mutans UA150 in complex with Gentiobiose 6-phosphate. | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-O-phosphono-beta-D-glucopyranose-(1-6)-beta-D-glucopyranose, 6-phospho-beta-D-Glucosidase, ... | | 著者 | Tan, K, Michalska, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-08-21 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.291 Å) | | 主引用文献 | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
4F79
 
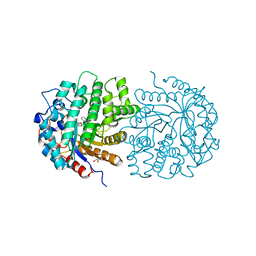 | | The crystal structure of 6-phospho-beta-glucosidase mutant (E375Q) in complex with Salicin 6-phosphate | | 分子名称: | 2-(hydroxymethyl)phenyl 6-O-phosphono-beta-D-glucopyranoside, GLYCEROL, Putative phospho-beta-glucosidase | | 著者 | Tan, K, Michalska, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-05-15 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
