2XIM
 
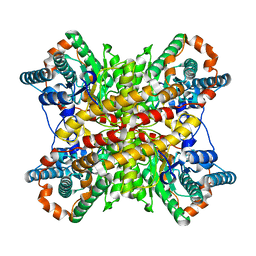 | | ARGININE RESIDUES AS STABILIZING ELEMENTS IN PROTEINS | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, Xylitol | | Authors: | Mrabet, N.T, Van Denbroek, A, Van Den Brande, I, Stanssens, P, Laroche, Y, Lambeir, A.-M, Matthyssens, G, Jenkins, J, Chiadmi, M, Vantilbeurgh, H, Rey, F, Janin, J, Quax, W.J, Lasters, I, Demaeyer, M, Wodak, S.J. | | Deposit date: | 1991-05-29 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Arginine residues as stabilizing elements in proteins.
Biochemistry, 31, 1992
|
|
3XIM
 
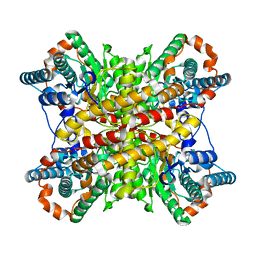 | | ARGININE RESIDUES AS STABILIZING ELEMENTS IN PROTEINS | | Descriptor: | COBALT (II) ION, D-XYLOSE ISOMERASE, sorbitol | | Authors: | Mrabet, N.T, Van Denbroek, A, Van Den Brande, I, Stanssens, P, Laroche, Y, Lambeir, A.-M, Matthyssens, G, Jenkins, J, Chiadmi, M, Vantilbeurgh, H, Rey, F, Janin, J, Quax, W.J, Lasters, I, Demaeyer, M, Wodak, S.J. | | Deposit date: | 1991-05-29 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Arginine residues as stabilizing elements in proteins.
Biochemistry, 31, 1992
|
|
1UCN
 
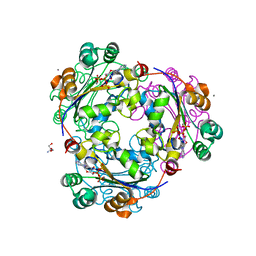 | | X-ray structure of human nucleoside diphosphate kinase A complexed with ADP at 2 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Chen, Y, Gallois-Montbrun, S, Schneider, B, Veron, M, Morera, S, Deville-Bonne, D, Janin, J. | | Deposit date: | 2003-04-16 | | Release date: | 2003-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nucleotide Binding to Nucleoside Diphosphate Kinases: X-ray Structure of Human NDPK-A in Complex with ADP and Comparison to Protein Kinases
J.Mol.Biol., 332, 2003
|
|
1T0I
 
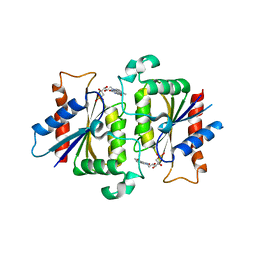 | | YLR011wp, a Saccharomyces cerevisiae NA(D)PH-dependent FMN reductase | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, YLR011wp | | Authors: | Liger, D, Graille, M, Zhou, C.-Z, Leulliot, N, Quevillon-Cheruel, S, Blondeau, K, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2004-04-09 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Characterization of Yeast YLR011wp, an Enzyme with NAD(P)H-FMN and Ferric Iron Reductase Activities
J.Biol.Chem., 279, 2004
|
|
1B4S
 
 | | STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE H122G MUTANT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE, ... | | Authors: | Meyer, P, Janin, J. | | Deposit date: | 1998-12-28 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleophilic activation by positioning in phosphoryl transfer catalyzed by nucleoside diphosphate kinase.
Biochemistry, 38, 1999
|
|
1BUX
 
 | | 3'-PHOSPHORYLATED NUCLEOTIDES BINDING TO NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Xu, Y, Schneider, B, Deville-Bonne, D, Veron, M, Janin, J. | | Deposit date: | 1998-09-07 | | Release date: | 1999-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3'-Phosphorylated nucleotides are tight binding inhibitors of nucleoside diphosphate kinase activity.
J.Biol.Chem., 273, 1998
|
|
2C5Q
 
 | | Crystal structure of yeast YER010Cp | | Descriptor: | 1,2-ETHANEDIOL, RRAA-LIKE PROTEIN YER010C | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Graille, M, Schiltz, M, Blondeau, K, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Yeast Yer010Cp, a Knotable Member of the Rraa Protein Family.
Protein Sci., 14, 2005
|
|
1XIM
 
 | | ARGININE RESIDUES AS STABILIZING ELEMENTS IN PROTEINS | | Descriptor: | COBALT (II) ION, D-XYLOSE ISOMERASE, Xylitol | | Authors: | Mrabet, N.T, Van Denbroek, A, Van Den Brande, I, Stanssens, P, Laroche, Y, Lambeir, A.-M, Matthyssens, G, Jenkins, J, Chiadmi, M, Vantilbeurgh, H, Rey, F, Janin, J, Quax, W.J, Lasters, I, Demaeyer, M, Wodak, S.J. | | Deposit date: | 1991-05-29 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arginine residues as stabilizing elements in proteins.
Biochemistry, 31, 1992
|
|
1KDN
 
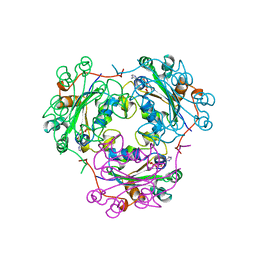 | | STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Cherfils, J, Xu, Y.W, Morera, S, Janin, J. | | Deposit date: | 1996-09-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AlF3 mimics the transition state of protein phosphorylation in the crystal structure of nucleoside diphosphate kinase and MgADP.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1EHW
 
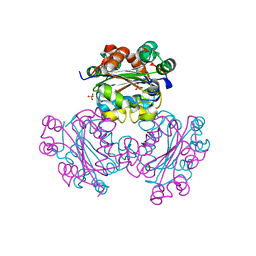 | | HUMAN NUCLEOSIDE DIPHOSPHATE KINASE 4 | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE, SULFATE ION | | Authors: | Milon, L, Meyer, P, Chiadmi, M, Munier, A, Johansson, M, Karlsson, A, Lascu, I, Capeau, J, Janin, J, Lacombe, M.-L. | | Deposit date: | 2000-02-23 | | Release date: | 2000-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase.
J.Biol.Chem., 275, 2000
|
|
1F6T
 
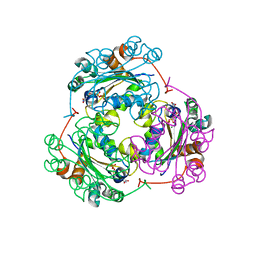 | | STRUCTURE OF THE NUCLEOSIDE DIPHOSPHATE KINASE/ALPHA-BORANO(RP)-TDP.MG COMPLEX | | Descriptor: | 2*-DEOXY-THYMIDINE-5*-ALPHA BORANO DIPHOSPHATE (ISOMER RP), MAGNESIUM ION, PROTEIN (NUCLEOSIDE DIPHOSPHATE KINASE) | | Authors: | Guerreiro, C, Boretto, J, Janin, J, Veron, M, Canard, B, Schneider, B, Sarfati, S, Deville-Bonne, D. | | Deposit date: | 2000-06-23 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for activation of alpha-boranophosphate nucleotide analogues targeting drug-resistant reverse transcriptase.
EMBO J., 19, 2000
|
|
1F3F
 
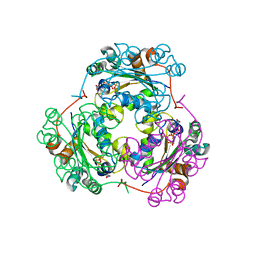 | | STRUCTURE OF THE H122G NUCLEOSIDE DIPHOSPHATE KINASE / D4T-TRIPHOSPHATE.MG COMPLEX | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-DIPHOSPHATE, 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Meyer, P, Schneider, B, Sarfati, S, Deville-Bonne, D, Guerreiro, C, Boretto, J, Janin, J, Veron, M, Canard, B. | | Deposit date: | 2000-06-02 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for activation of alpha-boranophosphate nucleotide analogues targeting drug-resistant reverse transcriptase.
EMBO J., 19, 2000
|
|
1NDC
 
 | |
1XE7
 
 | | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly roll fold | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GUANINE, ... | | Authors: | Zhou, C.-Z, Meyer, P, Quevillon-Cheruel, S, Li de La Sierra-Gallay, I, Collinet, B, Graille, M, Leulliot, N, Sorel, I, Janin, J, Van Tilbeurgh, H. | | Deposit date: | 2004-09-09 | | Release date: | 2005-01-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly-roll fold
Protein Sci., 14, 2005
|
|
1XE8
 
 | | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly roll fold. | | Descriptor: | ADENINE, CITRIC ACID, GLYCEROL, ... | | Authors: | Zhou, C.-Z, Meyer, P, Quevillon-Cheruel, S, Li de La Sierra-Gallay, I, Collinet, B, Graille, M, Leulliot, N, Sorel, I, Janin, J, Van Tilbeurgh, H. | | Deposit date: | 2004-09-09 | | Release date: | 2005-01-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly-roll fold
Protein Sci., 14, 2005
|
|
1YM5
 
 | | Crystal structure of YHI9, the yeast member of the phenazine biosynthesis PhzF enzyme superfamily. | | Descriptor: | Hypothetical 32.6 kDa protein in DAP2-SLT2 intergenic region | | Authors: | Liger, D, Quevillon-Cheruel, S, Sorel, I, Bremang, M, Blondeau, K, Aboulfath, I, Janin, J, Van Tilbeurgh, H, Leulliot, N, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2005-01-20 | | Release date: | 2005-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of YHI9, the yeast member of the phenazine biosynthesis PhzF enzyme superfamily
Proteins, 60, 2005
|
|
1YCD
 
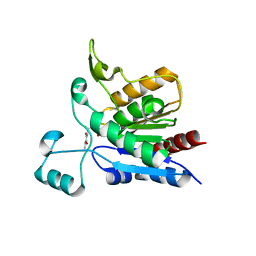 | | Crystal structure of yeast FSH1/YHR049W, a member of the serine hydrolase family | | Descriptor: | 2-HYDROXY-4,5-DIOXOHEPTYL HYDROGEN PHOSPHONATE, Hypothetical 27.3 kDa protein in AAP1-SMF2 intergenic region | | Authors: | Leulliot, N, Graille, M, Coste, F, Quevillon-Cheruel, S, Janin, J, van Tilbeurgh, H, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2004-12-22 | | Release date: | 2005-05-10 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of yeast YHR049W/FSH1, a member of the serine hydrolase family.
Protein Sci., 14, 2005
|
|
1K44
 
 | |
1XTZ
 
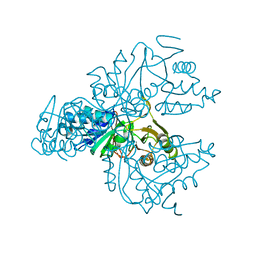 | | Crystal structure of the S. cerevisiae D-ribose-5-phosphate isomerase: comparison with the archeal and bacterial enzymes | | Descriptor: | Ribose-5-phosphate isomerase | | Authors: | Graille, M, Meyer, P, Leulliot, N, Sorel, I, Janin, J, van Tilbeurgh, H, Quevillon-Cheruel, S. | | Deposit date: | 2004-10-25 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the S. cerevisiae D-ribose-5-phosphate isomerase: comparison with the archaeal and bacterial enzymes
Biochimie, 87, 2005
|
|
2BKW
 
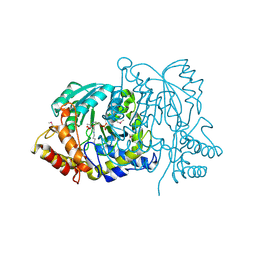 | | Yeast alanine:glyoxylate aminotransferase YFL030w | | Descriptor: | ALANINE-GLYOXYLATE AMINOTRANSFERASE 1, GLYOXYLIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Meyer, P, Liger, D, Leulliot, N, Quevillon-Cheruel, S, Zhou, C.Z, Borel, F, Ferrer, J.L, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2005-02-21 | | Release date: | 2005-11-02 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure and Confirmation of the Alanine:Glyoxylate Aminotransferase Activity of the Yfl030W Yeast Protein
Biochimie, 87, 2005
|
|
1G6Y
 
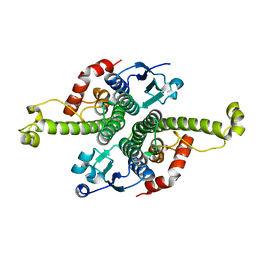 | | CRYSTAL STRUCTURE OF THE GLOBULAR REGION OF THE PRION PROTEIN URE2 FROM YEAST SACCHAROMYCES CEREVISIAE | | Descriptor: | URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Janin, J, Melki, R, Morera, S. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the globular region of the prion protein Ure2 from the yeast Saccharomyces cerevisiae.
Structure, 9, 2001
|
|
1HHQ
 
 | | Role of active site resiude Lys16 in Nucleoside Diphosphate Kinase | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE, SULFATE ION | | Authors: | Schneider, B, Babolat, M, Xu, Y.W, Janin, J, Veron, M, Deville-Bonne, D. | | Deposit date: | 2000-12-26 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of Phosphoryl Transfer by Nucleoside Diphosphate Kinase Ph-Dependence and Role of Active Site Lys16 and Tyr56 Residues
Eur.J.Biochem., 268, 2001
|
|
1HLW
 
 | | STRUCTURE OF THE H122A MUTANT OF THE NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Admiraal, S.J, Meyer, P, Schneider, B, Deville-Bonne, D, Janin, J, Herschlag, D. | | Deposit date: | 2000-12-04 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical rescue of phosphoryl transfer in a cavity mutant: a cautionary tale for site-directed mutagenesis.
Biochemistry, 40, 2001
|
|
1G6W
 
 | | CRYSTAL STRUCTURE OF THE GLOBULAR REGION OF THE PRION PROTEIN URE2 FROM THE YEAST SACCAROMYCES CEREVISIAE | | Descriptor: | URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Janin, J, Melki, R, Morera, S. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the globular region of the prion protein Ure2 from the yeast Saccharomyces cerevisiae.
Structure, 9, 2001
|
|
1HIY
 
 | | Binding of nucleotides to NDP kinase | | Descriptor: | 3'-DEOXY 3'-AMINO ADENOSINE-5'-DIPHOSPHATE, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Cervoni, L, Lascu, I, Xu, Y, Gonin, P, Morr, M, Merouani, M, Janin, J, Giartoso, A. | | Deposit date: | 2001-01-05 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of Nucleotides to Nucleoside Diphosphate Kinase: A Calorimetric Study.
Biochemistry, 40, 2001
|
|
