2PQU
 
 | |
2PY9
 
 | |
2P2R
 
 | | Crystal structure of the third KH domain of human Poly(C)-Binding Protein-2 in complex with C-rich strand of human telomeric DNA | | Descriptor: | 6-AMINOPYRIMIDIN-2(1H)-ONE, C-rich strand of human telomeric DNA, Poly(rC)-binding protein 2 | | Authors: | James, T.L, Stroud, R.M, Du, Z, Fenn, S, Tjhen, R, Lee, J.K. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the third KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.6 A resolution.
Nucleic Acids Res., 35, 2007
|
|
1B10
 
 | | SOLUTION NMR STRUCTURE OF RECOMBINANT SYRIAN HAMSTER PRION PROTEIN RPRP(90-231) , 25 STRUCTURES | | Descriptor: | PROTEIN (PRION PROTEIN) | | Authors: | James, T.L, Liu, H, Ulyanov, N.B, Farr-Jones, S. | | Deposit date: | 1998-11-25 | | Release date: | 1998-12-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
219D
 
 | |
28SP
 
 | |
28SR
 
 | |
1D42
 
 | |
2AXY
 
 | | Crystal Structure of KH1 domain of human Poly(C)-binding protein-2 with C-rich strand of human telomeric DNA | | Descriptor: | C-rich strand of human telomeric dna, Poly(rC)-binding protein 2 | | Authors: | Du, Z, Lee, J.K, Tjhen, R.J, Li, S, Stroud, R.M, James, T.L. | | Deposit date: | 2005-09-06 | | Release date: | 2005-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the First KH Domain of Human Poly(C)-binding Protein-2 in Complex with a C-rich Strand of Human Telomeric DNA at 1.7 A
J.Biol.Chem., 280, 2005
|
|
3C4B
 
 | | Structure of RNaseIIIb and dsRNA binding domains of mouse Dicer | | Descriptor: | Endoribonuclease Dicer | | Authors: | Lee, J.K, Du, Z, Tjhen, R.J, Stroud, R.M, James, T.L. | | Deposit date: | 2008-01-29 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and biochemical insights into the dicing mechanism of mouse Dicer: A conserved lysine is critical for dsRNA cleavage.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C4T
 
 | | Structure of RNaseIIIb and dsRNA binding domains of mouse Dicer | | Descriptor: | CADMIUM ION, Endoribonuclease Dicer | | Authors: | Lee, J.K, Du, Z, Tjhen, R.J, Stroud, R.M, James, T.L. | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into the dicing mechanism of mouse Dicer: A conserved lysine is critical for dsRNA cleavage.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
142D
 
 | |
141D
 
 | |
140D
 
 | | SOLUTION STRUCTURE OF A CONSERVED DNA SEQUENCE FROM THE HIV-1 GENOME: RESTRAINED MOLECULAR DYNAMICS SIMULATION WITH DISTANCE AND TORSION ANGLE RESTRAINTS DERIVED FROM TWO-DIMENSIONAL NMR SPECTRA | | Descriptor: | DNA (5'-D(*AP*GP*CP*TP*TP*GP*CP*CP*TP*TP*GP*AP*G)-3'), DNA (5'-D(*CP*TP*CP*AP*AP*GP*GP*CP*AP*AP*GP*CP*T)-3') | | Authors: | Mujeeb, A, Kerwin, S.M, Kenyon, G.L, James, T.L. | | Deposit date: | 1993-09-24 | | Release date: | 1994-04-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a conserved DNA sequence from the HIV-1 genome: restrained molecular dynamics simulation with distance and torsion angle restraints derived from two-dimensional NMR spectra.
Biochemistry, 32, 1993
|
|
1CQL
 
 | |
1D70
 
 | |
1R7W
 
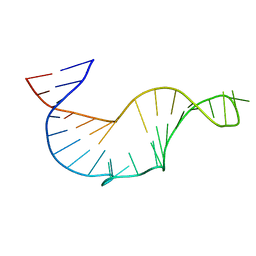 | | NMR STRUCTURE OF THE R(GGAGGACAUCCCUCACGGGUGACCGUGGUCCUCC), DOMAIN IV STEM-LOOP B OF ENTEROVIRAL IRES WITH AUCCCU BULGE | | Descriptor: | 34-MER | | Authors: | Du, Z, Ulyanov, N.B, Yu, J, James, T.L. | | Deposit date: | 2003-10-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Loop B RNAs from the Stem-Loop IV Domain of the Enterovirus Internal Ribosome Entry Site: A Single C to U Substitution Drastically Changes the Shape and Flexibility of RNA(,).
Biochemistry, 43, 2004
|
|
1R7Z
 
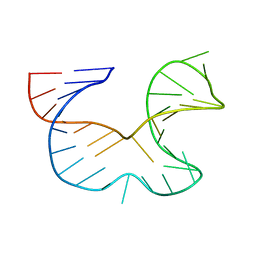 | | NMR STRUCTURE OF THE R(GGAGGACAUUCCUCACGGGUGACCGUGGUCCUCC), DOMAIN IV STEM-LOOP B OF ENTEROVIRAL IRES WITH AUUCCU BULGE | | Descriptor: | 34-MER | | Authors: | Du, Z, Ulyanov, N.B, Yu, J, James, T.L. | | Deposit date: | 2003-10-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Loop B RNAs from the Stem-Loop IV Domain of the Enterovirus Internal Ribosome Entry Site: A Single C to U Substitution Drastically Changes the Shape and Flexibility of RNA(,).
Biochemistry, 43, 2004
|
|
1BAU
 
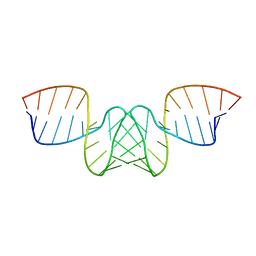 | | NMR STRUCTURE OF THE DIMER INITIATION COMPLEX OF HIV-1 GENOMIC RNA, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SL1 RNA DIMER | | Authors: | Mujeeb, A, Clever, J.L, Billeci, T.M, James, T.L, Parslow, T.G. | | Deposit date: | 1998-04-18 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the dimer initiation complex of HIV-1 genomic RNA.
Nat.Struct.Biol., 5, 1998
|
|
1CQ5
 
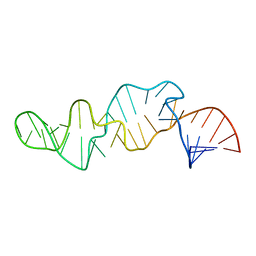 | | NMR STRUCTURE OF SRP RNA DOMAIN IV | | Descriptor: | SRP RNA DOMAIN IV | | Authors: | Schmitz, U, James, T.L, Behrens, S, Freymann, D.M, Lukavsky, P, Walter, P. | | Deposit date: | 1999-08-05 | | Release date: | 1999-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the phylogenetically most conserved domain of SRP RNA.
RNA, 5, 1999
|
|
2GM0
 
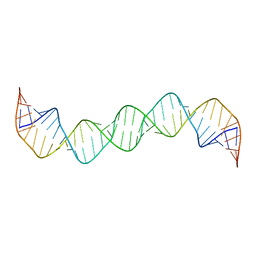 | | Linear dimer of stemloop SL1 from HIV-1 | | Descriptor: | RNA (35-MER) | | Authors: | Ulyanov, N.B, Mujeeb, A, Du, Z, Tonelli, M, Parslow, T.G, James, T.L. | | Deposit date: | 2006-04-05 | | Release date: | 2006-04-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Full-length Linear Dimer of Stem-Loop-1 RNA in the HIV-1 Dimer Initiation Site.
J.Biol.Chem., 281, 2006
|
|
1SLS
 
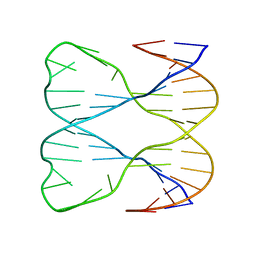 | | IMMOBILE SLIPPED-LOOP STRUCTURE (SLS) OF DNA HOMODIMER IN SOLUTION, NMR, 9 STRUCTURES | | Descriptor: | OLIGODEOXYRIBONUCLEOTIDE | | Authors: | Ulyanov, N.B, Ivanov, V.I, Minyat, E.E, Khomyakova, E.B, Petrova, M.V, Lesiak, K, James, T.L. | | Deposit date: | 1997-09-30 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A pseudosquare knot structure of DNA in solution.
Biochemistry, 37, 1998
|
|
1SKP
 
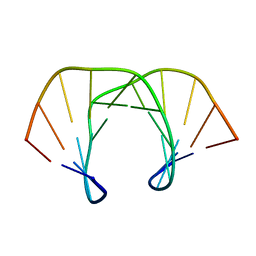 | | NMR STRUCTURE OF D(GCATATGATAG)(DOT)D(CTATCATATGC): A CONSENSUS SEQUENCE FOR PROMOTERS RECOGNIZED BY SIGMA-K RNA POLYMERASE, 4 STRUCTURES | | Descriptor: | SIGMA-K RNA POLYMERASE CONSENSUS SEQUENCE | | Authors: | Tonelli, M, Ragg, E, Bianucci, A.M, Lesiak, K, James, T.L. | | Deposit date: | 1998-05-20 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of d(GCATATGATAG). d(CTATCATATGC): a consensus sequence for promoters recognized by sigma K RNA polymerase.
Biochemistry, 37, 1998
|
|
1ROQ
 
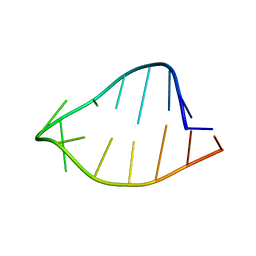 | |
1TXS
 
 | | STEM-LOOP D OF THE CLOVERLEAF DOMAIN OF ENTEROVIRAL 5'UTR RNA | | Descriptor: | Enteroviral 5'-UTR | | Authors: | Du, Z, Yu, J, Ulyanov, N.B, Andino, R, James, T.L. | | Deposit date: | 2004-07-06 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Consensus Stem-Loop D RNA Domain that Plays Important Roles in Regulating Translation and Replication in Enteroviruses and Rhinoviruses
Biochemistry, 43, 2004
|
|
