4CW4
 
 | |
4CSY
 
 | | E-selectin lectin, EGF-like and two SCR domains complexed with Sialyl Lewis X | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, E-SELECTIN, ... | | Authors: | Preston, R.C, Jakob, R.P, Binder, F.P.C, Sager, C.P, Ernst, B, Maier, T. | | Deposit date: | 2014-03-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | E-Selectin Ligand Complexes Adopt an Extended High-Affinity Conformation.
J.Mol.Cell.Biol., 8, 2016
|
|
4CSS
 
 | | Crystal structure of FimH in complex with a sulfonamide biphenyl alpha D-mannoside | | Descriptor: | 4'-(alpha-D-Mannopyranosyloxy)-biphenyl-4-methyl sulfonamide, GLYCEROL, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
4C16
 
 | | E-selectin lectin, EGF-like and two SCR domains complexed with glycomimetic antagonist | | Descriptor: | (1R,2R,3S)-3-methylcyclohexane-1,2-diol, (S)-CYCLOHEXYL LACTIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Preston, R.C, Jakob, R.P, Binder, F.P.C, Sager, C.P, Ernst, B, Maier, T. | | Deposit date: | 2013-08-09 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | E-Selectin Ligand Complexes Adopt an Extended High-Affinity Conformation.
J.Mol.Cell.Biol., 8, 2016
|
|
2KGJ
 
 | |
8B7S
 
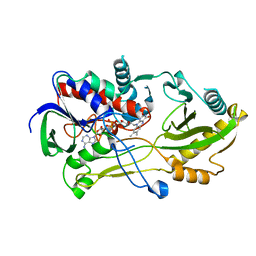 | | Crystal structure of the Chloramphenicol-inactivating oxidoreductase from Novosphingobium sp | | Descriptor: | Chloramphenicol-inactivating oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, L, Toplak, M, Saleem-Batcha, R, Hoeing, L, Jakob, R.P, Jehmlich, N, von Bergen, M, Maier, T, Teufel, R. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial Dehydrogenases Facilitate Oxidative Inactivation and Bioremediation of Chloramphenicol.
Chembiochem, 24, 2023
|
|
6FIJ
 
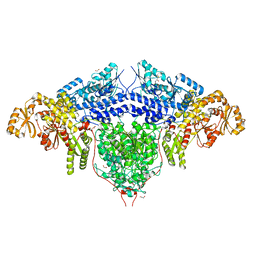 | | Structure of the loading/condensing region (SAT-KS-MAT) of the cercosporin fungal non-reducing polyketide synthase (NR-PKS) CTB1 | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, ... | | Authors: | Herbst, D.A, Jakob, R.P, Townsend, C.A, Maier, T. | | Deposit date: | 2018-01-18 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | The structural organization of substrate loading in iterative polyketide synthases.
Nat. Chem. Biol., 14, 2018
|
|
6FIK
 
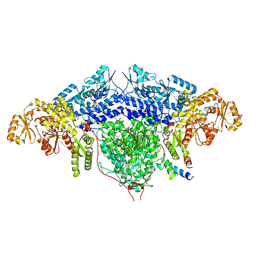 | | ACP2 crosslinked to the KS of the loading/condensing region of the CTB1 PKS | | Descriptor: | Polyketide synthase | | Authors: | Herbst, D.A, Huitt-Roehl, C.R, Jakob, R.P, Townsend, C.A, Maier, T. | | Deposit date: | 2018-01-18 | | Release date: | 2018-03-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | The structural organization of substrate loading in iterative polyketide synthases.
Nat. Chem. Biol., 14, 2018
|
|
4CST
 
 | | Crystal structure of FimH in complex with 3'-Chloro-4'-(alpha-D-mannopyranosyloxy)-biphenyl-4-carbonitrile | | Descriptor: | 3'-chloro-4'-(alpha-D-mannopyranosyloxy)biphenyl-4-carbonitrile, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
6FN6
 
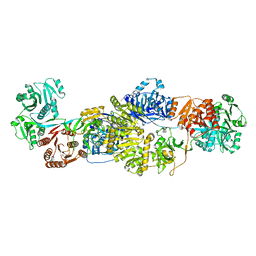 | | Modifying region (DH-ER-KR) of an insect fatty acid synthase (FAS) | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Fatty acid synthase 1, ... | | Authors: | Benning, F.M.C, Bukhari, H.S.T, Jakob, R.P, Maier, T. | | Deposit date: | 2018-02-02 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modifying region (DH-ER-KR) of an insect fatty acid synthase (FAS)
To Be Published
|
|
4X5Q
 
 | | Crystal structure of FimH in complex with 5-nitro-indolinylphenyl alpha-D-mannopyranoside | | Descriptor: | 4-(5-nitro-1H-indol-1-yl)phenyl alpha-D-mannopyranoside, Protein FimH | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
4X5R
 
 | | Crystal structure of FimH in complex with a squaryl-phenyl alpha-D-mannopyranoside derivative | | Descriptor: | 2-chloro-4-{[2-(4-methylpiperazin-1-yl)-3,4-dioxocyclobut-1-en-1-yl]amino}phenyl alpha-D-mannopyranoside, Protein FimH, SULFATE ION | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
4Z37
 
 | |
4Z42
 
 | | Crystal structure of urease from Yersinia enterocolitica | | Descriptor: | NICKEL (II) ION, Urease subunit alpha, Urease subunit beta, ... | | Authors: | Studer, G, Jakob, R.P, Mahi, M.A, Wiesand, U, Schwede, T, Maier, T. | | Deposit date: | 2015-04-01 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure of urease from Yersinia enterocolitica
To Be Published
|
|
4X50
 
 | | Crystal structure of FimH in complex with biphenyl alpha-D-mannopyranoside | | Descriptor: | Protein FimH, biphenyl-4-yl alpha-D-mannopyranoside | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-04 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
4X5P
 
 | | Crystal structure of FimH in complex with a benzoyl-amidophenyl alpha-D-mannopyranoside | | Descriptor: | 4-{[3-chloro-4-(alpha-D-mannopyranosyloxy)phenyl]carbamoyl}benzoic acid, Protein FimH | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.997 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
