1HVU
 
 | |
4RY6
 
 | |
4RY4
 
 | |
4RY7
 
 | |
4RY5
 
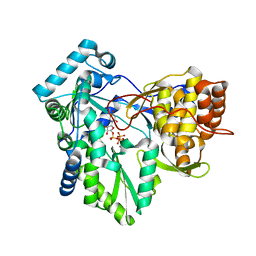 | | C-terminal mutant (W550N) of HCV/J4 RNA polymerase | | 分子名称: | HCV J4 RNA polymerase (NS5B), MANGANESE (II) ION, URIDINE 5'-TRIPHOSPHATE | | 著者 | Jaeger, J, Cherry, A, Dennis, C. | | 登録日 | 2014-12-13 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Hydrophobic and Charged Residues in the C-Terminal Arm of Hepatitis C Virus RNA-Dependent RNA Polymerase Regulate Initiation and Elongation.
J.Virol., 89, 2015
|
|
1ASM
 
 | |
1ASN
 
 | |
1ASL
 
 | |
1AIA
 
 | |
1AIC
 
 | |
1AIB
 
 | |
1NB4
 
 | |
3HVT
 
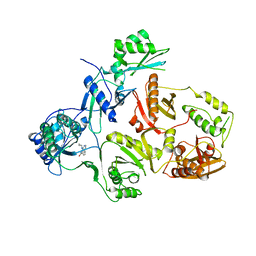 | | STRUCTURAL BASIS OF ASYMMETRY IN THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE HETERODIMER | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | 著者 | Steitz, T.A, Smerdon, S.J, Jaeger, J, Wang, J, Kohlstaedt, L.A, Chirino, A.J, Friedman, J.M, Rice, P.A. | | 登録日 | 1994-07-25 | | 公開日 | 1994-10-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.Usa, 91, 1994
|
|
1JN3
 
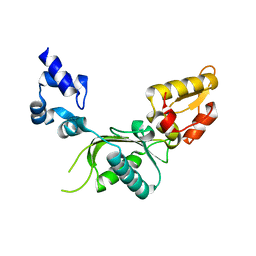 | |
6SY4
 
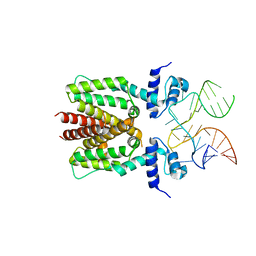 | | TetR in complex with the TetR-binding RNA-aptamer K1 | | 分子名称: | TetR-binding aptamer K1 (43-MER), Tetracycline repressor protein class B from transposon Tn10 | | 著者 | Grau, F.C, Muller, Y.A, Suess, B, Groher, F, Jaeger, J. | | 登録日 | 2019-09-27 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.695 Å) | | 主引用文献 | The complex formed between a synthetic RNA aptamer and the transcription repressor TetR is a structural and functional twin of the operator DNA-TetR regulator complex.
Nucleic Acids Res., 48, 2020
|
|
6SY6
 
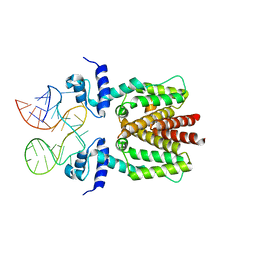 | | TetR in complex with the TetR-binding RNA-aptamer K2 | | 分子名称: | RNA (36-MER), Tetracycline repressor protein class B from transposon Tn10 | | 著者 | Grau, F.C, Muller, Y.A, Suess, B, Groher, F, Jaeger, J. | | 登録日 | 2019-09-27 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The complex formed between a synthetic RNA aptamer and the transcription repressor TetR is a structural and functional twin of the operator DNA-TetR regulator complex.
Nucleic Acids Res., 48, 2020
|
|
3TB8
 
 | |
1DYU
 
 | | The active site base controls cofactor reactivity in Escherichia coli amine oxidase: X-ray crystallographic studies with mutational variants. | | 分子名称: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | 著者 | Murray, J.M, Wilmot, C.M, Saysell, C.G, Jaeger, J, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | 登録日 | 2000-02-08 | | 公開日 | 2000-02-29 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | The Active Site Base Controls Cofactor Reactivity in Escherichia Coli Amine Oxidase : X-Ray Crystallographicstudies with Mutational Variants
Biochemistry, 38, 1999
|
|
3V7J
 
 | |
3V7K
 
 | |
3V7L
 
 | | Apo Structure of Rat DNA polymerase beta K72E variant | | 分子名称: | CHLORIDE ION, DNA polymerase beta, SODIUM ION, ... | | 著者 | Rangarajan, S, Jaeger, J. | | 登録日 | 2011-12-21 | | 公開日 | 2013-01-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Crystallographic studies of K72E mutant DNA polymerase explain loss of lyase function and reveal changes in the overall conformational state of the polymerase domain
To be Published
|
|
3V72
 
 | |
3UXO
 
 | |
3UXP
 
 | |
3UXN
 
 | |
