5JMS
 
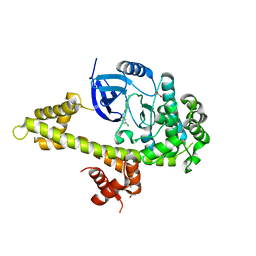 | | Crystal structure of TgCDPK1 bound to CGP060476 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calmodulin-domain protein kinase 1, ... | | Authors: | El Bakkouri, M, Walker, J.R, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Lovato, D.V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-04-29 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of TgCDPK1bount to CGP060476
To Be Published
|
|
5JO5
 
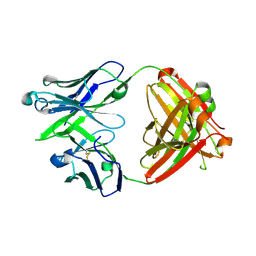 | |
5IZD
 
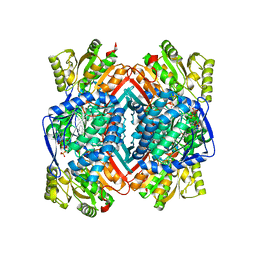 | |
4X55
 
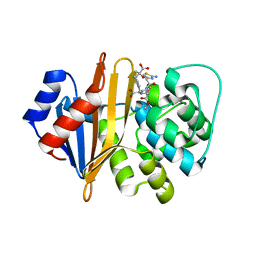 | | Structure of the class D Beta-Lactamase OXA-225 K82D in Acyl-Enzyme Complex with Ceftazidime | | Descriptor: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase OXA-225 | | Authors: | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
4X53
 
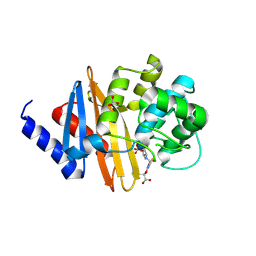 | | Structure of the class D Beta-Lactamase OXA-160 V130D in Acyl-Enzyme Complex with Aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, BICARBONATE ION, Class D beta-lactamase OXA-160 | | Authors: | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
4X56
 
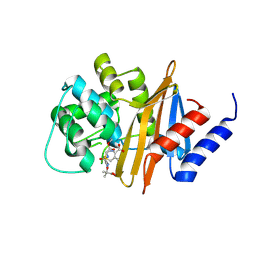 | | Structure of the class D Beta-Lactamase OXA-160 V130D in Acyl-Enzyme Complex with Ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Class D beta-lactamase OXA-160 | | Authors: | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
1U71
 
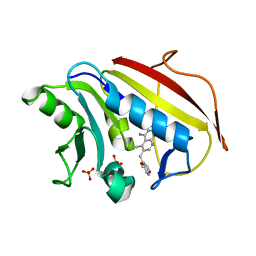 | | Understanding the Role of Leu22 Variants in Methotrexate Resistance: Comparison of Wild-type and Leu22Arg Variant Mouse and Human Dihydrofolate Reductase Ternary Crystal Complexes with Methotrexate and NADPH | | Descriptor: | 6-(2,5-DIMETHOXY-BENZYL)-5-METHYL-PYRIDO[2,3-D]PYRIMIDINE-2,4-DIAMINE, Dihydrofolate reductase, SULFATE ION | | Authors: | Cody, V, Luft, J.R, Pangborn, W. | | Deposit date: | 2004-08-02 | | Release date: | 2005-02-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Understanding the role of Leu22 variants in methotrexate resistance: comparison of wild-type and Leu22Arg variant mouse and human dihydrofolate reductase ternary crystal complexes with methotrexate and NADPH.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1UD7
 
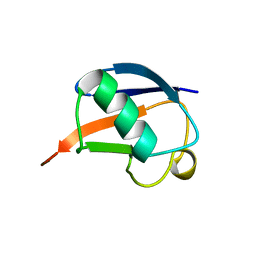 | | SOLUTION STRUCTURE OF THE DESIGNED HYDROPHOBIC CORE MUTANT OF UBIQUITIN, 1D7 | | Descriptor: | PROTEIN (UBIQUITIN CORE MUTANT 1D7) | | Authors: | Johnson, E.C, Lazar, G.A, Desjarlais, J.R, Handel, T.M. | | Deposit date: | 1999-04-07 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of a designed hydrophobic core variant of ubiquitin.
Structure Fold.Des., 7, 1999
|
|
1U35
 
 | | Crystal structure of the nucleosome core particle containing the histone domain of macroH2A | | Descriptor: | H2A histone family, Hist1h4i protein, Histone H3.1, ... | | Authors: | Chakravarthy, S, Gundimella, S.K, Caron, C, Perche, P.Y, Pehrson, J.R, Khochbin, S, Luger, K. | | Deposit date: | 2004-07-20 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural characterization of the histone variant macroH2A.
Mol.Cell.Biol., 25, 2005
|
|
1U00
 
 | | HscA substrate binding domain complexed with the IscU recognition peptide ELPPVKIHC | | Descriptor: | Chaperone protein hscA, IscU recognition peptide | | Authors: | Cupp-Vickery, J.R, Peterson, J.C, Ta, D.T, Vickery, L.E. | | Deposit date: | 2004-07-12 | | Release date: | 2004-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Molecular Chaperone HscA Substrate Binding Domain Complexed with the IscU Recognition Peptide ELPPVKIHC.
J.Mol.Biol., 342, 2004
|
|
1U8E
 
 | | HUMAN DIPEPTIDYL PEPTIDASE IV/CD26 MUTANT Y547F | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bjelke, J.R, Christensen, J, Branner, S, Wagtmann, N, Olsen, C, Kanstrup, A.B, Rasmussen, H.B. | | Deposit date: | 2004-08-05 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tyrosine 547 constitutes an essential part of the catalytic mechanism of dipeptidyl peptidase IV
J.Biol.Chem., 279, 2004
|
|
1U0I
 
 | | IAAL-E3/K3 heterodimer | | Descriptor: | IAAL-E3, IAAL-K3 | | Authors: | Lindhout, D.A, Litowski, J.R, Mercier, P, Hodges, R.S, Sykes, B.D. | | Deposit date: | 2004-07-13 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a highly stable de novo heterodimeric coiled-coil
Biopolymers, 75, 2004
|
|
1U11
 
 | | PurE (N5-carboxyaminoimidazole Ribonucleotide Mutase) from the acidophile Acetobacter aceti | | Descriptor: | CITRIC ACID, PurE (N5-carboxyaminoimidazole Ribonucleotide Mutase) | | Authors: | Settembre, E.C, Chittuluru, J.R, Mill, C.P, Kappock, T.J, Ealick, S.E. | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acidophilic adaptations in the structure of Acetobacter aceti N5-carboxyaminoimidazole ribonucleotide mutase (PurE).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1DLR
 
 | | METHOTREXATE-RESISTANT VARIANTS OF HUMAN DIHYDROFOLATE REDUCTASE WITH SUBSTITUTION OF LEUCINE 22: KINETICS, CRYSTALLOGRAPHY AND POTENTIAL AS SELECTABLE MARKERS | | Descriptor: | 6-(2,5-DIMETHOXY-BENZYL)-5-METHYL-PYRIDO[2,3-D]PYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W. | | Deposit date: | 1995-01-25 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Methotrexate-resistant variants of human dihydrofolate reductase with substitutions of leucine 22. Kinetics, crystallography, and potential as selectable markers.
J.Biol.Chem., 270, 1995
|
|
1DLS
 
 | | METHOTREXATE-RESISTANT VARIANTS OF HUMAN DIHYDROFOLATE REDUCTASE WITH SUBSTITUTION OF LEUCINE 22: KINETICS, CRYSTALLOGRAPHY AND POTENTIAL AS SELECTABLE MARKERS | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W. | | Deposit date: | 1995-01-25 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Methotrexate-resistant variants of human dihydrofolate reductase with substitutions of leucine 22. Kinetics, crystallography, and potential as selectable markers.
J.Biol.Chem., 270, 1995
|
|
7VD5
 
 | | Structure of C2S2M2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Nagao, R, Kato, K, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2021-09-06 | | Release date: | 2022-03-02 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for different types of hetero-tetrameric light-harvesting complexes in a diatom PSII-FCPII supercomplex
Nat Commun, 13, 2022
|
|
7Y3F
 
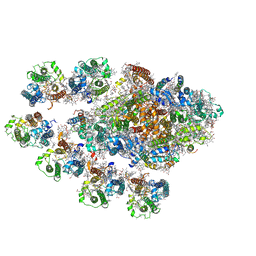 | | Structure of the Anabaena PSI-monomer-IsiA supercomplex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Nagao, R, Kato, K, Hamaguchi, T, Kawakami, K, Yonekura, K, Shen, J.R. | | Deposit date: | 2022-06-10 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of a monomeric photosystem I core associated with iron-stress-induced-A proteins from Anabaena sp. PCC 7120.
Nat Commun, 14, 2023
|
|
7XQP
 
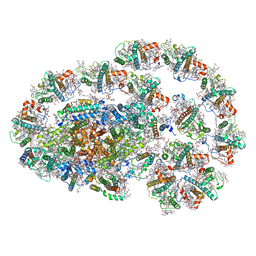 | | PSI-LHCI-LHCII-Lhcb9 supercomplex of Physcomitrella patens | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Zhang, S, Tang, K.L, Li, X.Y, Wang, W.D, Yan, Q.J, Shen, L.L, Kuang, T.Y, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural insights into a unique PSI-LHCI-LHCII-Lhcb9 supercomplex from moss Physcomitrium patens.
Nat.Plants, 9, 2023
|
|
7W2V
 
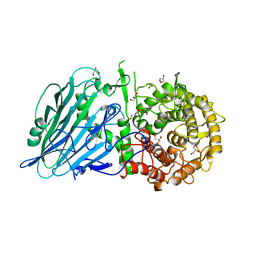 | | Crystal structure of TxGH116 R786A mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7W2S
 
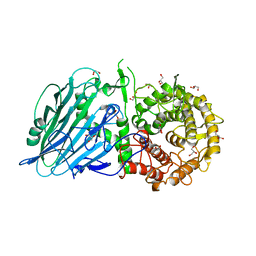 | | Crystal structure of TxGH116 E730A mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7W2W
 
 | | Crystal structure of TxGH116 R786K mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7W2T
 
 | | Crystal structure of TxGH116 E730Q mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Glucosylceramidase, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7W2X
 
 | | Crystal structure of TxGH116 R786K mutant from Thermoanaerobacterium xylanolyticum | | Descriptor: | CALCIUM ION, GLYCEROL, Glucosylceramidase, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
5M1Y
 
 | |
6GWK
 
 | | The crystal structure of Hfq from Caulobacter crescentus | | Descriptor: | RNA-binding protein Hfq | | Authors: | Santiago-Frangos, A, Frohlich, K.S, Jeliazkov, J.R, Gray, J.R, Luisi, B.F, Woodson, S.A, Hardwick, S.W. | | Deposit date: | 2018-06-25 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Caulobacter crescentusHfq structure reveals a conserved mechanism of RNA annealing regulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
