1CPB
 
 | |
2JBJ
 
 | | membrane-bound glutamate carboxypeptidase II (GCPII) in complex with 2-PMPA (2-phosphonoMethyl-pentanedioic acid) | | Descriptor: | (2S)-2-(PHOSPHONOMETHYL)PENTANEDIOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mesters, J.R, Henning, K, Hilgenfeld, R. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Human Glutamate Carboxypeptidase II Inhibition: Structures of Gcpii in Complex with Two Potent Inhibitors, Quisqualate and 2-Pmpa.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6XE4
 
 | | BTK Fluorocyclopropyl amide inhibitor, Compound 25 | | Descriptor: | (1S,2S)-N-[2'-(6-tert-butyl-8-fluoro-1-oxophthalazin-2(1H)-yl)-3'-(hydroxymethyl)-1-methyl-6-oxo[1,6-dihydro[3,4'-bipyridine]]-5-yl]-2-fluorocyclopropane-1-carboxamide, SULFATE ION, Tyrosine-protein kinase BTK | | Authors: | Kiefer, J.R, Crawford, J.J, Lee, W, Eigenbrot, C, Yu, C. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Stereochemical Differences in Fluorocyclopropyl Amides Enable Tuning of Btk Inhibition and Off-Target Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
6X7M
 
 | | LnmK in complex with 2-nitronate-propionyl-oxa(dethia)-CoA | | Descriptor: | Bifunctional methylmalonyl-CoA:ACP acyltransferase/decarboxylase, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Stunkard, L.M, Kick, B.J, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structures of LnmK, a Bifunctional Acyltransferase/Decarboxylase, with Substrate Analogues Reveal the Basis for Selectivity and Stereospecificity.
Biochemistry, 60, 2021
|
|
6WJR
 
 | |
6WK2
 
 | | SETD3 mutant (N255V) in Complex with an Actin Peptide with His73 Replaced with Methionine | | Descriptor: | 1,2-ETHANEDIOL, Actin, cytoplasmic 2, ... | | Authors: | Dai, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Characterization of SETD3 methyltransferase-mediated protein methionine methylation.
J.Biol.Chem., 295, 2020
|
|
6WE9
 
 | | YTH domain of human YTHDC1 with 11mer ssDNA Containing N6mA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*GP*(6MA)P*CP*TP*CP*TP*G)-3'), GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biochemical and structural basis for YTH domain of human YTHDC1 binding to methylated adenine in DNA.
Nucleic Acids Res., 48, 2020
|
|
6WF6
 
 | | Streptomyces coelicolor methylmalonyl-CoA epimerase | | Descriptor: | COBALT (II) ION, DI(HYDROXYETHYL)ETHER, Methylmalonyl-CoA epimerase | | Authors: | Stunkard, L.M, Benjamin, A.B, Bower, J.B, Huth, T.J, Lohman, J.R. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Substrate Enolate Intermediate and Mimic Captured in the Active Site of Streptomyces coelicolor Methylmalonyl-CoA Epimerase.
Chembiochem, 23, 2022
|
|
6WFH
 
 | | Streptomyces coelicolor methylmalonyl-CoA epimerase substrate complex | | Descriptor: | (3S,5R,9R,19E)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9,19-tetrahydroxy-8,8,20-trimethyl-10,14-dioxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphahenicos-19-en-21-oic acid 3,5-dioxide (non-preferred name), CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Stunkard, L.M, Benjamin, A.B, Bower, J.B, Huth, T.J, Lohman, J.R. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Substrate Enolate Intermediate and Mimic Captured in the Active Site of Streptomyces coelicolor Methylmalonyl-CoA Epimerase.
Chembiochem, 23, 2022
|
|
6YGN
 
 | | Titin kinase and its flanking domains | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Titin | | Authors: | Fleming, J.R, Franke, B, Bogomolovas, J, Mayans, O. | | Deposit date: | 2020-03-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Titin kinase ubiquitination aligns autophagy receptors with mechanical signals in the sarcomere.
Embo Rep., 22, 2021
|
|
6X7O
 
 | | LnmK in complex with 2-sulfonate-propionyl-CoA | | Descriptor: | (2R)-sulfonatepropionyl-CoA, (2S)-sulfonatepropionyl-CoA, Bifunctional methylmalonyl-CoA:ACP acyltransferase/decarboxylase, ... | | Authors: | Stunkard, L.M, Kick, B.J, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of LnmK, a Bifunctional Acyltransferase/Decarboxylase, with Substrate Analogues Reveal the Basis for Selectivity and Stereospecificity.
Biochemistry, 60, 2021
|
|
1E6C
 
 | | K15M MUTANT OF SHIKIMATE KINASE FROM ERWINIA CHRYSANTHEMI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Maclean, J, Krell, T, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2000-08-10 | | Release date: | 2001-06-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and X-Ray Crystallographic Studies on Shikimate Kinase: The Important Structural Role of the P-Loop Lysine
Protein Sci., 10, 2001
|
|
6X6P
 
 | | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Herrera, N.G, Morano, N.C, Celikgil, A, Georgiev, G.I, Malonis, R, Lee, J.H, Tong, K, Vergnolle, O, Massimi, A, Yen, L.Y, Noble, A.J, Kopylov, M, Bonanno, J.B, Garrett-Thompson, S.C, Hayes, D.B, Brenowitz, M, Garforth, S.J, Eng, E.T, Lai, J.R, Almo, S.C. | | Deposit date: | 2020-05-28 | | Release date: | 2020-06-10 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis.
Biorxiv, 2020
|
|
6X7N
 
 | | LnmK in complex with 2-nitronate-propionyl-amino(dethia)-CoA | | Descriptor: | Bifunctional methylmalonyl-CoA:ACP acyltransferase/decarboxylase, SULFATE ION, [1-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethylamino]-1-oxidanylidene-propan-2-ylidene]-bis(oxidanidyl)azanium | | Authors: | Stunkard, L.M, Kick, B.J, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structures of LnmK, a Bifunctional Acyltransferase/Decarboxylase, with Substrate Analogues Reveal the Basis for Selectivity and Stereospecificity.
Biochemistry, 60, 2021
|
|
2JAC
 
 | |
2FLC
 
 | |
6YXE
 
 | | Structure of the Trim69 RING domain | | Descriptor: | E3 ubiquitin-protein ligase TRIM69, ZINC ION | | Authors: | Keown, J.R, Goldstone, D.C. | | Deposit date: | 2020-05-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The RING domain of TRIM69 promotes higher-order assembly.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2FL7
 
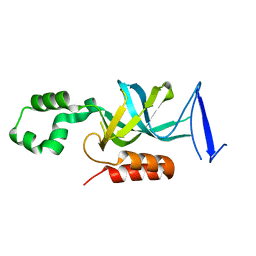 | | S. cerevisiae Sir3 BAH domain | | Descriptor: | Regulatory protein SIR3 | | Authors: | Keck, J.L, Hou, Z, Daner, J.R, Fox, C.A. | | Deposit date: | 2006-01-05 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the Sir3 protein bromo adjacent homology (BAH) domain from S. cerevisiae at 1.95 A resolution.
Protein Sci., 15, 2006
|
|
2FKC
 
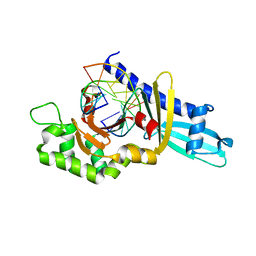 | |
1BLS
 
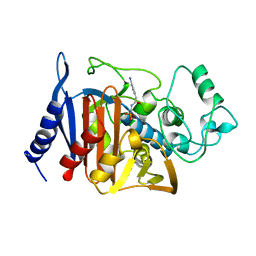 | |
2FKH
 
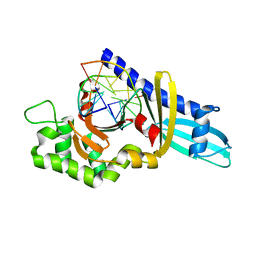 | |
7SMH
 
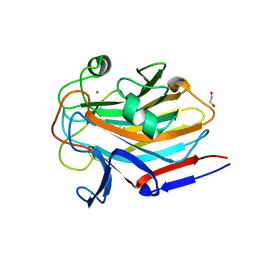 | | Structure of SASG A-domain (residues 163-419) from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Surface protein G | | Authors: | Atkin, K.E, Whelan, F, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
7SP2
 
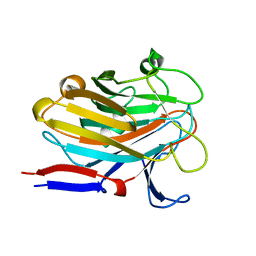 | | Structure of PLS A-domain (residues 391-656; 513-518 deletion mutant) from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Plasmin Sensitive Protein Pls | | Authors: | Clark, L, Whelan, F, Atkin, K.E, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of PLS A-domain (residues 391-65) from Staphylococcus aureus
Not Published
|
|
7TN0
 
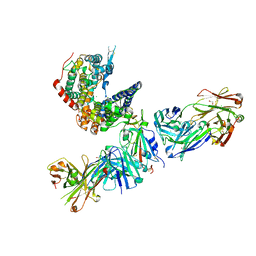 | | SARS-CoV-2 Omicron RBD in complex with human ACE2 and S304 Fab and S309 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | McCallum, M, Czudnochowski, N, Nix, J.C, Croll, T.I, SSGCID, Dillen, J.R, Snell, G, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement.
Science, 375, 2022
|
|
7TT7
 
 | | BamABCDE bound to substrate EspP in the barrelized EspP/continuous open BamA state | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
