3CDC
 
 | |
3LQ6
 
 | |
3CDY
 
 | | AL-09 H87Y, immunoglobulin light chain variable domain | | Descriptor: | IMMUNOGLOBULIN LIGHT CHAIN | | Authors: | Baden, E.M, Randles, E.G, Aboagye, A.K, Thompson, J.R, Ramirez-Alvarado, M. | | Deposit date: | 2008-02-27 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural insights into the role of mutations in amyloidogenesis.
J.Biol.Chem., 283, 2008
|
|
3CB9
 
 | |
3GNF
 
 | | P1 Crystal structure of the N-terminal R1-R7 of murine MVP | | Descriptor: | Major vault protein | | Authors: | Querol-Audi, J, Casanas, A, Uson, I, Luque, D, Caston, J.R, Fita, I, Verdaguer, N. | | Deposit date: | 2009-03-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP
Embo J., 28, 2009
|
|
3CDF
 
 | |
5GTI
 
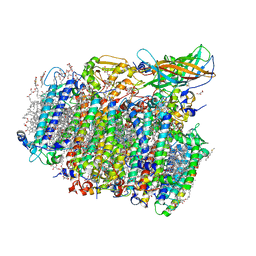 | | Native XFEL structure of photosystem II (two flash dataset) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-08-20 | | Release date: | 2017-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
5H2F
 
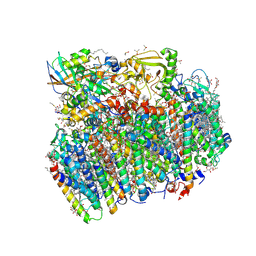 | | Crystal structure of the PsbM-deletion mutant of photosystem II | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Uto, S, Kawakami, K, Umena, Y, Iwai, M, Ikeuchi, M, Shen, J.R, Kamiya, N. | | Deposit date: | 2016-10-15 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutual relationships between structural and functional changes in a PsbM-deletion mutant of photosystem II.
Faraday Discuss., 198, 2017
|
|
3CUX
 
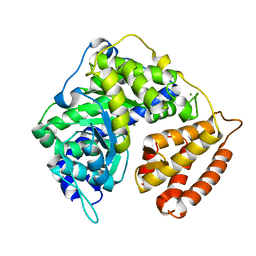 | |
3LIJ
 
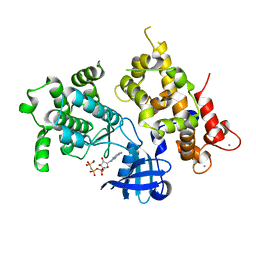 | | Crystal structure of full length CpCDPK3 (cgd5_820) in complex with Ca2+ and AMPPNP | | Descriptor: | CALCIUM ION, Calcium/calmodulin dependent protein kinase with a kinase domain and 4 calmodulin like EF hands, MAGNESIUM ION, ... | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Walker, J.R, Sullivan, H, Lin, Y.-H, Mackenzie, F, Kozieradzki, I, Cossar, D, Schapira, M, Senisterra, G, Vedadi, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-25 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of full length CpCDPK3 (cgd5_820) in complex with Ca2+ and AMPPNP
To be Published
|
|
3HI1
 
 | | Structure of HIV-1 gp120 (core with V3) in Complex with CD4-Binding-Site Antibody F105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F105 Heavy Chain, F105 Light Chain, ... | | Authors: | Kwon, Y.D, Chen, L, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z, Zhang, M.-Y, Arthos, J, Burton, D.R, Dimitrov, D, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-05-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
5GTH
 
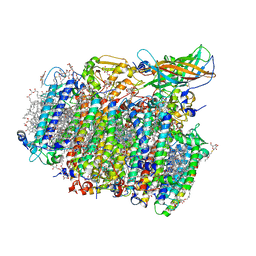 | | Native XFEL structure of photosystem II (dark dataset) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-08-20 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
3HIL
 
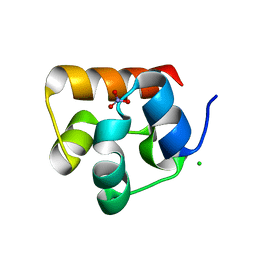 | | SAM Domain of Human Ephrin Type-A Receptor 1 (EphA1) | | Descriptor: | CHLORIDE ION, Ephrin type-A receptor 1, NITRATE ION | | Authors: | Walker, J.R, Yermekbayeva, L, Butler-Cole, C, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-20 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SAM Domain of Human Ephrin Type-A Receptor 1 (EphA1).
To be Published
|
|
4IUW
 
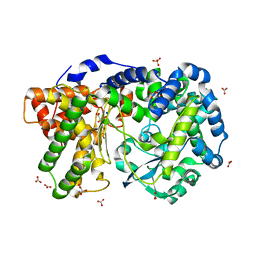 | | Crystal structure of PEPO from Lactobacillus rhamnosis HN001 (DR20) | | Descriptor: | CARBONATE ION, CITRIC ACID, Neutral endopeptidase, ... | | Authors: | Anderson, B.F, Knapp, K.M, Holland, R, Norris, G.E, Christensson, C, Bratt, H, Collins, L.J, Coolbear, T, Lubbers, M.W, Toole, P.W.O, Reid, J.R, Jameson, G.B. | | Deposit date: | 2013-01-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of PEPO from Lactobacillus rhamnosis HN001 (DR20)
TO BE PUBLISHED
|
|
3LFC
 
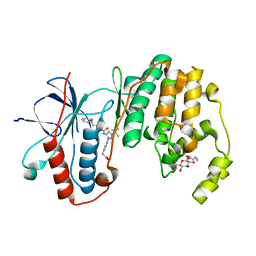 | | Human p38 MAP Kinase in Complex with RL99 | | Descriptor: | (4-{5-[({4-[2-(benzyloxy)ethyl]-1,3-thiazol-2-yl}carbamoyl)amino]-3-tert-butyl-1H-pyrazol-1-yl}phenyl)acetic acid, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-01-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha
To be Published
|
|
3BAY
 
 | | N298S Variant of Human Pancreatic Alpha-Amylase in Complex with Nitrate and Acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACARBOSE DERIVED PENTASACCHARIDE, CALCIUM ION, ... | | Authors: | Fredriksen, J.R, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-11-08 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
3LJZ
 
 | | Crystal Structure of Human MMP-13 complexed with an Amino-2-indanol compound | | Descriptor: | (2R)-2-[4-(1,3-benzodioxol-5-yl)benzyl]-N~4~-hydroxy-N~1~-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]butanediamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Shieh, H.-S, Kiefer, J.R. | | Deposit date: | 2010-01-26 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure analysis reveals the flexibility of the ADAMTS-5 active site.
Protein Sci., 20, 2011
|
|
4J0B
 
 | | Structure of mitochondrial Hsp90 (TRAP1) with ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, COBALT (II) ION, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-30 | | Release date: | 2014-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
3AZR
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3B6T
 
 | | Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) T686A Mutant in Complex with Quisqualate at 2.1 Resolution | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Cho, Y, Lolis, E, Howe, J.R. | | Deposit date: | 2007-10-29 | | Release date: | 2008-02-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and single-channel results indicate that the rates of ligand binding domain closing and opening directly impact AMPA receptor gating.
J.Neurosci., 28, 2008
|
|
3LC9
 
 | |
4IVG
 
 | | Crystal structure of mitochondrial Hsp90 (TRAP1) NTD-Middle domain dimer with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-22 | | Release date: | 2014-01-22 | | Last modified: | 2014-08-27 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
3FZU
 
 | | IgG1 Fab characterized by H/D exchange | | Descriptor: | immunoglobulin IgG1 Fab, heavy chain, light chain | | Authors: | Arndt, J, Houde, D, Domeier, W, Berkowitz, S, Engen, J.R. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of IgG1 conformation and conformational dynamics by hydrogen/deuterium exchange mass spectrometry.
Anal Chem, 81, 2009
|
|
3LFF
 
 | | Human p38 MAP Kinase in Complex with RL166 | | Descriptor: | (4-{3-tert-butyl-5-[(1,3-thiazol-2-ylcarbamoyl)amino]-1H-pyrazol-1-yl}phenyl)acetic acid, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-01-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha
To be Published
|
|
4IYN
 
 | | Structure of mitochondrial Hsp90 (TRAP1) with ADP-ALF4- | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
