2RJC
 
 | | Crystal structure of L3MBTL1 protein in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
3QJ6
 
 | | The crystal structure of PWWP domain of human Hepatoma-derived growth factor 2 in complex with H3K79me3 peptide | | Descriptor: | H3K79me3, Hepatoma-derived growth factor-related protein 2, SULFATE ION, ... | | Authors: | Zeng, H, Amaya, M.F, Tempel, W, Walker, J.R, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-28 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
2RGL
 
 | | Rice BGlu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2007-10-04 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into Rice BGlu1 beta-Glucosidase Oligosaccharide Hydrolysis and Transglycosylation
J.Mol.Biol., 377, 2008
|
|
2RGM
 
 | | Rice BGlu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucosidase, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2007-10-04 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insights into Rice BGlu1 beta-Glucosidase Oligosaccharide Hydrolysis and Transglycosylation
J.Mol.Biol., 377, 2008
|
|
2RJF
 
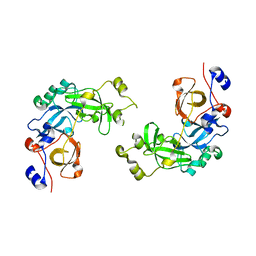 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 12-30), orthorhombic form I | | Descriptor: | Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
7KS0
 
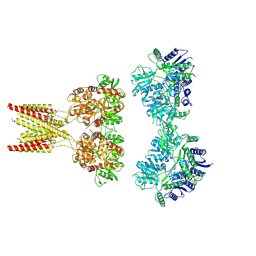 | | GluK2/K5 with 6-Cyano-7-nitroquinoxaline-2,3-dione (CNQX) | | Descriptor: | Glutamate receptor ionotropic, kainate 2, kainate 5,Green fluorescent protein chimera | | Authors: | Khanra, N, Brown, P.M.G.E, Perozzo, A.M, Bowie, D, Meyerson, J.R. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Architecture and structural dynamics of the heteromeric GluK2/K5 kainate receptor.
Elife, 10, 2021
|
|
7KS3
 
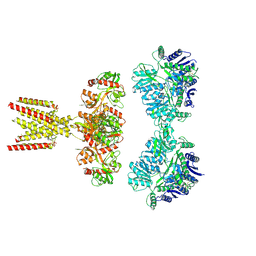 | | GluK2/K5 with L-Glu | | Descriptor: | Glutamate receptor ionotropic, kainate 2, kainate 5,Green fluorescent protein chimera | | Authors: | Khanra, N, Brown, P.M.G.E, Perozzo, A.M, Bowie, D, Meyerson, J.R. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Architecture and structural dynamics of the heteromeric GluK2/K5 kainate receptor.
Elife, 10, 2021
|
|
2RJE
 
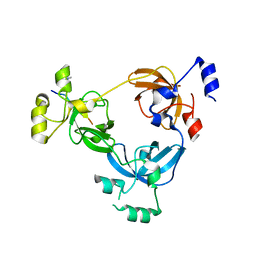 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 17-25), orthorhombic form II | | Descriptor: | CHLORIDE ION, Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
3J6D
 
 | | Model of the PrgH-PrgK periplasmic rings | | Descriptor: | Pathogenicity 1 island effector protein, Protein PrgH | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2014-02-14 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | The Modular Structure of the Inner-Membrane Ring Component PrgK Facilitates Assembly of the Type III Secretion System Basal Body.
Structure, 23, 2015
|
|
7SZ2
 
 | | Mouse PARP13/ZAP ZnF5-WWE1-WWE2 bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ayanath Kuttiyatveetil, J.R, Pascal, J.M. | | Deposit date: | 2021-11-25 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and functional analysis of the ZnF5-WWE1-WWE2 region of PARP13/ZAP define a distinctive mode of engaging poly(ADP-ribose).
Cell Rep, 41, 2022
|
|
6NPK
 
 | | Structure of the TM domain | | Descriptor: | Solute carrier family 12 (sodium/potassium/chloride transporter), member 2 | | Authors: | Feng, L, Liao, M.F, Orlando, B, Zhang, J.R. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-31 | | Last modified: | 2019-08-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and mechanism of the cation-chloride cotransporter NKCC1.
Nature, 572, 2019
|
|
5I3L
 
 | | DPF3b in complex with H3K14ac peptide | | Descriptor: | 1,2-ETHANEDIOL, H3K14ac peptide, SODIUM ION, ... | | Authors: | Tempel, W, Liu, Y, Walker, J.R, Zhao, A, Qin, S, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-10 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of DPF3b in complex with an acetylated histone peptide.
J.Struct.Biol., 195, 2016
|
|
6NQC
 
 | |
7SZ3
 
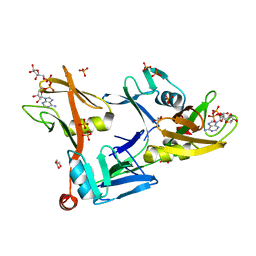 | | Mouse PARP13/ZAP ZnF5-WWE1-WWE2 bound to ADPr | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ayanath Kuttiyatveetil, J.R, Pascal, J.M. | | Deposit date: | 2021-11-25 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and functional analysis of the ZnF5-WWE1-WWE2 region of PARP13/ZAP define a distinctive mode of engaging poly(ADP-ribose).
Cell Rep, 41, 2022
|
|
6NPJ
 
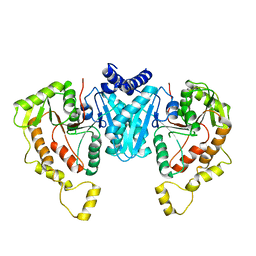 | | Structure of the NKCC1 CTD | | Descriptor: | Sodium-potassium-chloride cotransporter 1 | | Authors: | Feng, L, Liao, M.F, Orlando, B, Zhang, J.R. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and mechanism of the cation-chloride cotransporter NKCC1.
Nature, 572, 2019
|
|
7JYA
 
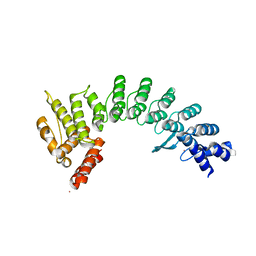 | | Crystal structure of E3 ligase in complex with peptide | | Descriptor: | ASN-ARG-ARG-ARG-ARG-TRP-ARG-GLU-ARG-GLN-ARG, Protein fem-1 homolog C, UNKNOWN ATOM OR ION | | Authors: | Yan, X, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Dong, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-30 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Molecular basis for ubiquitin ligase CRL2 FEM1C -mediated recognition of C-degron.
Nat.Chem.Biol., 17, 2021
|
|
6NPH
 
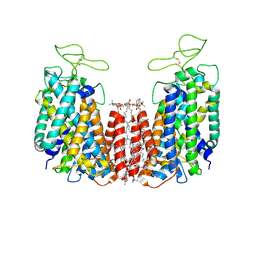 | | Structure of NKCC1 TM domain | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Feng, L, Liao, M.F, Orlando, B, Zhang, J.R. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-31 | | Last modified: | 2020-01-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and mechanism of the cation-chloride cotransporter NKCC1.
Nature, 572, 2019
|
|
7SVS
 
 | |
3WU2
 
 | | Crystal structure analysis of Photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Umena, Y, Kawakami, K, Shen, J.R, Kamiya, N. | | Deposit date: | 2014-04-21 | | Release date: | 2014-09-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 A
Nature, 473, 2011
|
|
3MY0
 
 | | Crystal structure of the ACVRL1 (ALK1) kinase domain bound to LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Serine/threonine-protein kinase receptor R3 | | Authors: | Chaikuad, A, Alfano, I, Cooper, C, Mahajan, P, Daga, N, Sanvitale, C, Fedorov, O, Petrie, K, Savitsky, P, Gileadi, O, Sethi, R, Krojer, T, Muniz, J.R.C, Pike, A.C.W, Vollmar, M, Carpenter, C.P, Ugochukwu, E, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A small molecule targeting ALK1 prevents Notch cooperativity and inhibits functional angiogenesis.
Angiogenesis, 18, 2015
|
|
3RQR
 
 | | Crystal structure of the RYR domain of the rabbit ryanodine receptor | | Descriptor: | (UNK)(UNK)(UNK)(UNK), Ryanodine receptor 1 | | Authors: | Nair, U.B, Li, W, Dong, A, Walker, J.R, Gramolini, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-28 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural determination of the phosphorylation domain of the ryanodine receptor.
Febs J., 279, 2012
|
|
6NPL
 
 | | Cryo-EM structure of NKCC1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Feng, L, Liao, M.F, Orlando, B, Zhang, J.R. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-31 | | Last modified: | 2019-08-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and mechanism of the cation-chloride cotransporter NKCC1.
Nature, 572, 2019
|
|
3MAO
 
 | | Crystal Structure of Human Methionine-R-Sulfoxide Reductase B1 (MsrB1) | | Descriptor: | FE (III) ION, MALONATE ION, Methionine-R-sulfoxide reductase B1, ... | | Authors: | Chaikuad, A, Shafqat, N, Yue, W.W, Savitsky, P, Krojer, T, Ugochukwu, E, Muniz, J.R.C, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of Human Methionine-R-Sulfoxide Reductase B1 (MsrB1)
To be Published
|
|
7SCY
 
 | | Nuc147 bound to single BRCT | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B type 1-J, ... | | Authors: | Muthurajan, U.M, Rudolph, J.R. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The BRCT domain of PARP1 binds intact DNA and mediates intrastrand transfer.
Mol.Cell, 81, 2021
|
|
3TYY
 
 | | Crystal Structure of Human Lamin-B1 Coil 2 Segment | | Descriptor: | Lamin-B1 | | Authors: | Lam, R, Xu, C, Bian, C.B, Mackenzie, F, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal structures of the coil 2B fragment and the globular tail domain of human lamin B1.
Febs Lett., 586, 2012
|
|
