4X55
 
 | | Structure of the class D Beta-Lactamase OXA-225 K82D in Acyl-Enzyme Complex with Ceftazidime | | 分子名称: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase OXA-225 | | 著者 | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | 登録日 | 2014-12-04 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.941 Å) | | 主引用文献 | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
3R1B
 
 | | Open crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | 分子名称: | (4-tert-butylphenyl)acetaldehyde, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | 著者 | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | 登録日 | 2011-03-09 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
3R1A
 
 | | Closed crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | 分子名称: | (4-tert-butylphenyl)acetaldehyde, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | 登録日 | 2011-03-09 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
1VRN
 
 | | PHOTOSYNTHETIC REACTION CENTER BLASTOCHLORIS VIRIDIS (ATCC) | | 分子名称: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | 著者 | Baxter, R.H.G, Seagle, B.-L, Norris, J.R. | | 登録日 | 2005-02-23 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Cryogenic structure of the photosynthetic reaction center of Blastochloris viridis in the light and dark.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1VJ3
 
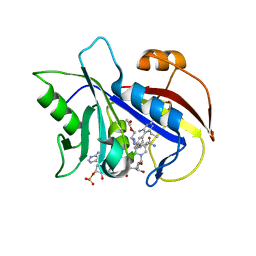 | | STRUCTURAL STUDIES ON BIO-ACTIVE COMPOUNDS. CRYSTAL STRUCTURE AND MOLECULAR MODELING STUDIES ON THE PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE COFACTOR COMPLEX WITH TAB, A HIGHLY SELECTIVE ANTIFOLATE. | | 分子名称: | ACETIC ACID N-[2-CHLORO-5-[6-ETHYL-2,4-DIAMINO-PYRIMID-5-YL]-PHENYL]-[BENZYL-TRIAZEN-3-YL]ETHYL ESTER, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Cody, V, Galitsky, N, Rak, D, Luft, J.R, Queener, S.F, Laughton, C.A, Malcolm, F.G. | | 登録日 | 2004-01-13 | | 公開日 | 2004-01-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural studies on bioactive compounds. 30. Crystal structure and molecular modeling studies on the Pneumocystis carinii dihydrofolate reductase cofactor complex with TAB, a highly selective antifolate.
Biochemistry, 39, 2000
|
|
4BDP
 
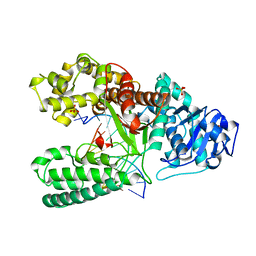 | | CRYSTAL STRUCTURE OF BACILLUS DNA POLYMERASE I FRAGMENT COMPLEXED TO 11 BASE PAIRS OF DUPLEX DNA AFTER ADDITION OF TWO DATP RESIDUES | | 分子名称: | DNA (5'-D(*GP*CP*AP*TP*CP*AP*TP*GP*CP*AP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*GP*CP*AP*TP*GP*AP*TP*GP*C)-3'), MAGNESIUM ION, ... | | 著者 | Kiefer, J.R, Mao, C, Beese, L.S. | | 登録日 | 1997-11-17 | | 公開日 | 1999-01-13 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal.
Nature, 391, 1998
|
|
1YF3
 
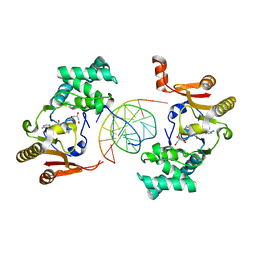 | | T4Dam in Complex with AdoHcy and 13-mer Oligonucleotide Making Non- and Semi-specific (~1/4) Contact | | 分子名称: | 5'-D(*AP*CP*CP*AP*TP*GP*AP*TP*CP*TP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*AP*GP*AP*TP*CP*AP*TP*GP*G)-3', DNA adenine methylase, ... | | 著者 | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | 登録日 | 2004-12-30 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
3MVR
 
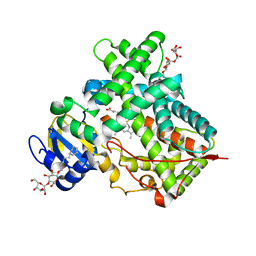 | |
1YFL
 
 | | T4Dam in Complex with Sinefungin and 16-mer Oligonucleotide Showing Semi-specific and Specific Contact and Flipped Base | | 分子名称: | 5'-D(P*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*GP*A)-3', DNA adenine methylase, SINEFUNGIN | | 著者 | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | 登録日 | 2005-01-03 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
6GWK
 
 | | The crystal structure of Hfq from Caulobacter crescentus | | 分子名称: | RNA-binding protein Hfq | | 著者 | Santiago-Frangos, A, Frohlich, K.S, Jeliazkov, J.R, Gray, J.R, Luisi, B.F, Woodson, S.A, Hardwick, S.W. | | 登録日 | 2018-06-25 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Caulobacter crescentusHfq structure reveals a conserved mechanism of RNA annealing regulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6H4C
 
 | | A polyamorous repressor: deciphering the evolutionary strategy used by the phage-inducible chromosomal islands to spread in nature. | | 分子名称: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, NICKEL (II) ION, ... | | 著者 | Ciges-Tomas, J.R, Alite, C, Bowring, J.Z, Donderis, J, Penades, J.R, Marina, A. | | 登録日 | 2018-07-20 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | The structure of a polygamous repressor reveals how phage-inducible chromosomal islands spread in nature.
Nat Commun, 10, 2019
|
|
1XWL
 
 | |
1YFJ
 
 | | T4Dam in Complex with AdoHcy and 15-mer Oligonucleotide Showing Semi-specific and Specific Contact | | 分子名称: | 5'-D(*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*G)-3', CALCIUM ION, CHLORIDE ION, ... | | 著者 | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | 登録日 | 2005-01-02 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
3KW4
 
 | | Crystal structure of cytochrome 2B4 in complex with the anti-platelet drug ticlopidine | | 分子名称: | 2-{[(3alpha,5alpha,7alpha,8alpha,10alpha,12alpha,17alpha)-3,12-bis{2-[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]ethoxy}cholan-7-yl]oxy}ethyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | 著者 | Gay, S.C, Maekawa, K, Roberts, A.G, Hong, W.-X, Zhang, Q, Stout, C.D, Halpert, J.R. | | 登録日 | 2009-11-30 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel.
Biochemistry, 49, 2010
|
|
1YPN
 
 | | REDUCED FORM HYDROXYMETHYLBILANE SYNTHASE (K59Q MUTANT) CRYSTAL STRUCTURE AFTER 2 HOURS IN A FLOW CELL DETERMINED BY TIME-RESOLVED LAUE DIFFRACTION | | 分子名称: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, HYDROXYMETHYLBILANE SYNTHASE | | 著者 | Helliwell, J.R, Nieh, Y.P, Raftery, J, Cassetta, A, Habash, J, Carr, P.D, Ursby, T, Wulff, M, Thompson, A.W, Niemann, A.C, Haedener, A. | | 登録日 | 1998-06-26 | | 公開日 | 1999-03-02 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Time-Resolved Structures of Hydroxymethylbilane Synthase (Lys59Gln Mutant) as It Isloaded with Substrate in the Crystal Determined by Laue Diffraction
J.Chem.Soc.,Faraday Trans., 94, 1998
|
|
3BDP
 
 | | DNA POLYMERASE I/DNA COMPLEX | | 分子名称: | DNA (5'-D(*AP*GP*CP*AP*TP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*GP*CP*2DT)-3'), PROTEIN (DNA POLYMERASE I), ... | | 著者 | Kiefer, J.R, Mao, C, Beese, L.S. | | 登録日 | 1997-11-17 | | 公開日 | 1999-01-13 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal.
Nature, 391, 1998
|
|
6L4U
 
 | | Structure of the PSI-FCPI supercomplex from diatom | | 分子名称: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | Nagao, R, Kato, K, Miyazaki, N, Akita, F, Shen, J.R. | | 登録日 | 2019-10-21 | | 公開日 | 2020-05-20 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Structure of photosystem I-light-harvesting supercomplex from a red-lineage diatom
Nat Commun, 2020
|
|
6L4T
 
 | | Structure of the peripheral FCPI from diatom | | 分子名称: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | Nagao, R, Kato, K, Miyazaki, N, Akita, F, Shen, J.R. | | 登録日 | 2019-10-21 | | 公開日 | 2020-05-20 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structure of photosystem I-light-harvesting supercomplex from a red-lineage diatom
Nat Commun, 2020
|
|
6LF1
 
 | | SeviL, a GM1b/asialo-GM1 binding lectin | | 分子名称: | CHLORIDE ION, SeviL | | 著者 | Kamata, K, Ozeki, Y, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2019-11-28 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The structure of SeviL, a GM1b/asialo-GM1 binding R-type lectin from the mussel Mytilisepta virgata.
Sci Rep, 10, 2020
|
|
6LF2
 
 | | SeviL bound to asialo-GM1 saccharide | | 分子名称: | SeviL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Kamata, K, Ozeki, Y, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2019-11-28 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The structure of SeviL, a GM1b/asialo-GM1 binding R-type lectin from the mussel Mytilisepta virgata.
Sci Rep, 10, 2020
|
|
5E6H
 
 | |
6MBP
 
 | | GLP Methyltransferase with Inhibitor EML741- P3121 Crystal Form | | 分子名称: | 1,2-ETHANEDIOL, 2-cyclohexyl-7-methoxy-N-[1-(propan-2-yl)piperidin-4-yl]-8-[3-(pyrrolidin-1-yl)propoxy]-3H-1,4-benzodiazepin-5-amine, Histone-lysine N-methyltransferase EHMT1, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2018-08-30 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.947 Å) | | 主引用文献 | Discovery of a Novel Chemotype of Histone Lysine Methyltransferase EHMT1/2 (GLP/G9a) Inhibitors: Rational Design, Synthesis, Biological Evaluation, and Co-crystal Structure.
J. Med. Chem., 62, 2019
|
|
5ISL
 
 | |
5IVV
 
 | |
5IW0
 
 | |
