4GSA
 
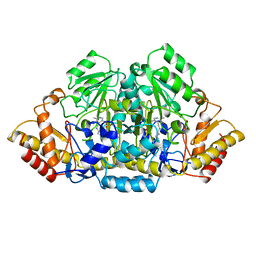 | |
6NP6
 
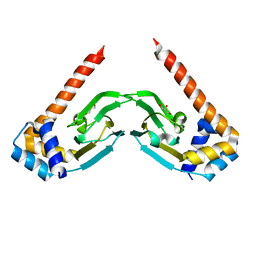 | | Crystal structure of the sensor domain of the transcriptional regulator HcpR from Porphyromonas Gingivalis | | Descriptor: | Crp/Fnr family transcriptional regulator, GLYCEROL | | Authors: | Musayev, F.N, Belvin, B.R, Escalante, C.R, Turner, J, Scarsdale, J.N, Lewis, J.P. | | Deposit date: | 2019-01-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nitrosative Stress Sensing in Porphyromonas gingivalis: Structure and Mechanisms of the Heme Binding Transcriptional Regulator HcpR.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1JS6
 
 | | Crystal Structure of DOPA decarboxylase | | Descriptor: | DOPA decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Burkhard, P, Dominici, P, Borri-Voltattorni, C, Jansonius, J.N, Malashkevich, V.N. | | Deposit date: | 2001-08-16 | | Release date: | 2001-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase.
Nat.Struct.Biol., 8, 2001
|
|
2WT1
 
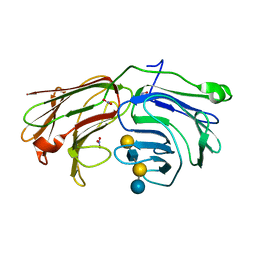 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre complexed with lacto-N-neo-tetraose | | Descriptor: | NITRATE ION, PUTATIVE FIBER PROTEIN, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
2WST
 
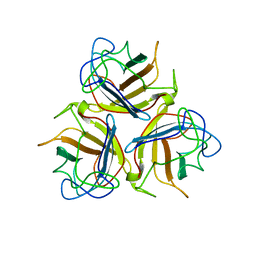 | | Head domain of porcine adenovirus type 4 NADC-1 isolate fibre | | Descriptor: | PUTATIVE FIBER PROTEIN | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
2WT0
 
 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre complexed with N-acetyl-lactosamine | | Descriptor: | NITRATE ION, PUTATIVE FIBER PROTEIN, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
2WSV
 
 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre complexed with lactose | | Descriptor: | NITRATE ION, PUTATIVE FIBER PROTEIN, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
2WT2
 
 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre complexed with tri(N-acetyl-lactosamine) | | Descriptor: | NITRATE ION, PUTATIVE FIBER PROTEIN, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
2WSU
 
 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre | | Descriptor: | GLYCEROL, NITRATE ION, PUTATIVE FIBER PROTEIN | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
5JP1
 
 | | Structure of Xanthomonas campestris effector protein XopD bound to tomato SUMO | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, MALONATE ION, Small ubiquitin-related modifier, ... | | Authors: | Pruneda, J.N, Komander, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Molecular Basis for Ubiquitin and Ubiquitin-like Specificities in Bacterial Effector Proteases.
Mol.Cell, 63, 2016
|
|
5KIS
 
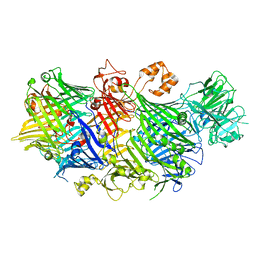 | | YenB/RHS2 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, RHS2, ... | | Authors: | Busby, J.N, Hurst, M.R.H, Lott, J.S. | | Deposit date: | 2016-06-16 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a new BC complex from Yersinia entomophaga
To Be Published
|
|
7UG6
 
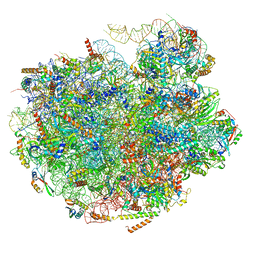 | | Cryo-EM structure of pre-60S ribosomal subunit, unmethylated G2922 | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Yelland, J.N, Bravo, J.P.K, Black, J.J.B, Taylor, D.W, Johnson, A.W. | | Deposit date: | 2022-03-24 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A single 2'-O-methylation of ribosomal RNA gates assembly of a functional ribosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7UYG
 
 | |
7UYH
 
 | |
5JP3
 
 | |
7AOY
 
 | |
6ZTQ
 
 | | Cryo-EM structure of respiratory complex I from Mus musculus inhibited by piericidin A at 3.0 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bridges, H.R, Blaza, J.N, Agip, A.N.A, Hirst, J. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-21 | | Last modified: | 2020-10-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of inhibitor-bound mammalian complex I.
Nat Commun, 11, 2020
|
|
6ZR2
 
 | | Cryo-EM structure of respiratory complex I in the active state from Mus musculus at 3.1 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bridges, H.R, Blaza, J.N, Agip, A.N.A, Hirst, J. | | Deposit date: | 2020-07-10 | | Release date: | 2020-10-21 | | Last modified: | 2020-10-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of inhibitor-bound mammalian complex I.
Nat Commun, 11, 2020
|
|
5LO9
 
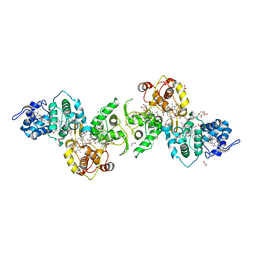 | | Thiosulfate dehydrogenase (TsdBA) from Marichromatium purpuratum - "as isolated" form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cytochrome C, ... | | Authors: | Brito, J.A, Kurth, J.M, Reuter, J, Flegler, A, Koch, T, Franke, T, Klein, E, Rowe, S, Butt, J.N, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2016-08-08 | | Release date: | 2016-10-12 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Electron Accepting Units of the Diheme Cytochrome c TsdA, a Bifunctional Thiosulfate Dehydrogenase/Tetrathionate Reductase.
J.Biol.Chem., 291, 2016
|
|
5M45
 
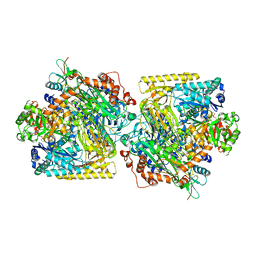 | | Structure of Acetone Carboxylase purified from Xanthobacter autotrophicus | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kabasakal, B.V, Wells, J.N, Nwaobi, B.C, Eilers, B.J, Peters, J.W, Murray, J.W. | | Deposit date: | 2016-10-18 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
2MCG
 
 | |
6PCV
 
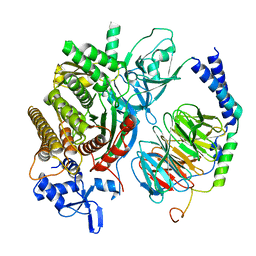 | | Single Particle Reconstruction of Phosphatidylinositol (3,4,5) trisphosphate-dependent Rac exchanger 1 bound to G protein beta gamma subunits | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Phosphatidylinositol (3,4,5) trisphosphate-dependent Rac exchanger 1 | | Authors: | Cash, J.N, Cianfrocco, M.A, Tesmer, J.J.G. | | Deposit date: | 2019-06-18 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-electron microscopy structure and analysis of the P-Rex1-G beta gamma signaling scaffold.
Sci Adv, 5, 2019
|
|
8C6P
 
 | | Fragment screening hit I bound to endothiapepsin | | Descriptor: | 4-[(2-azanyl-4-methyl-1,3-thiazol-5-yl)methyl]benzenecarbonitrile, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C6Q
 
 | | Fragment screening hit II bound to endothiapepsin | | Descriptor: | 3-(4-methylimidazol-1-yl)-5-(trifluoromethyl)aniline, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C71
 
 | | Pyrrolidine fragment 5b bound to endothiapepsin | | Descriptor: | (3~{R},4~{R})-4-(3,4-dihydro-1~{H}-isoquinolin-2-yl)pyrrolidin-3-ol, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
