4V57
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with spectinomycin and neomycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Shoji, S, Holton, J.M, Fredrick, K, Cate, J.H.D. | | Deposit date: | 2007-07-21 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A steric block in translation caused by the antibiotic spectinomycin.
Acs Chem.Biol., 2, 2007
|
|
4V4Q
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli at 3.5 A resolution. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schuwirth, B.S, Borovinskaya, M.A, Hau, C.W, Zhang, W, Vila-Sanjurjo, A, Holton, J.M, Cate, J.H.D. | | Deposit date: | 2005-08-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of the bacterial ribosome at 3.5 A resolution.
Science, 310, 2005
|
|
8JIX
 
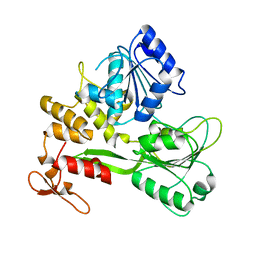 | |
8OJR
 
 | | Solution NMR Structure of Alpha-Synuclein 1-25 Peptide in 50% TFE. | | Descriptor: | Alpha-synuclein | | Authors: | Allen, S.G, Williams, C, Meade, R.M, Tang, T.M.S, Crump, M.P, Mason, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-08-09 | | Method: | SOLUTION NMR | | Cite: | An N-terminal alpha-Synuclein fragment binds lipid vesicles to modulate lipid induced aggregation
Cell Rep Phys Sci, 2023
|
|
8OH9
 
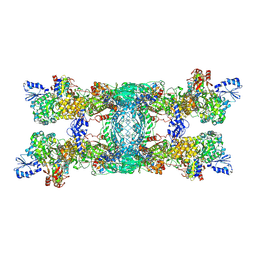 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 1) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Formate dehydrogenase-O, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
8OH5
 
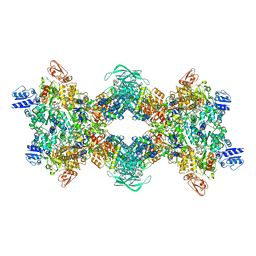 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 2) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
8OK0
 
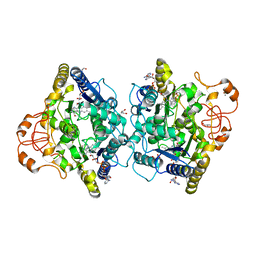 | | Crystal structure of human NQO1 in complex with the inhibitor PMSF | | Descriptor: | ACETYL GROUP, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Martin-Garcia, J.M, Grieco, A, Ruiz-Fresneda, M.A. | | Deposit date: | 2023-03-26 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural dynamics at the active site of the cancer-associated flavoenzyme NQO1 probed by chemical modification with PMSF.
Febs Lett., 597, 2023
|
|
8OGD
 
 | | Structure of zinc(II) double mutant human carbonic anhydrase II bound to thiocyanate | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, THIOCYANATE ION, ... | | Authors: | Silva, J.M, Cerofolini, L, Carvalho, A.L, Ravera, E, Fragai, M, Parigi, G, Macedo, A.L, Geraldes, C.F.G.C, Luchinat, C. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Elucidating the concentration-dependent effects of thiocyanate binding to carbonic anhydrase.
J.Inorg.Biochem., 244, 2023
|
|
8OGE
 
 | | Structure of cobalt(II) substituted double mutant human carbonic anhydrase II bound to thiocyanate | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, COBALT (II) ION, Carbonic anhydrase 2, ... | | Authors: | Silva, J.M, Cerofolini, L, Carvalho, A.L, Ravera, E, Fragai, M, Parigi, G, Macedo, A.L, Geraldes, C.F.G.C, Luchinat, C. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Elucidating the concentration-dependent effects of thiocyanate binding to carbonic anhydrase.
J.Inorg.Biochem., 244, 2023
|
|
8JJG
 
 | | Crystal structure of QW-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJI
 
 | | Crystal structure of QR-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JK0
 
 | | Crystal structure of QL-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJH
 
 | | Crystal structure of QH-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJX
 
 | | Crystal structure of QS-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJF
 
 | | Crystal structure of QE-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJZ
 
 | | Crystal structure of QQ-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJU
 
 | | Crystal structure of QD-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JK1
 
 | | Crystal structure of QA-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.067 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8OJS
 
 | | Crystal structure of the human IgD Fab - structure Fab1 | | Descriptor: | 1,2-ETHANEDIOL, Human IgD Fab heavy chain, Human IgD Fab light chain, ... | | Authors: | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
8OJU
 
 | | Crystal structure of the human IgD Fab - structure Fab3 | | Descriptor: | 1,2-ETHANEDIOL, Human IgD Fab heavy chain, Human IgD Fab light chain, ... | | Authors: | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
8OJV
 
 | | Crystal structure of the human IgD Fab - structure Fab4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
8OJT
 
 | | Crystal structure of the human IgD Fab - structure Fab2 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, ... | | Authors: | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
8JJY
 
 | | Crystal structure of QN-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJW
 
 | | Crystal structure of QG-hNTAQ1 C28S | | Descriptor: | MAGNESIUM ION, Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JK2
 
 | | Crystal structure of QF-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
