4ZMW
 
 | | Crystal structure of human P-cadherin (enc-X-dimer) | | Descriptor: | CALCIUM ION, Cadherin-3, NICKEL (II) ION, ... | | Authors: | Caaveiro, J.M.M, Kudo, S, Tsumoto, K. | | Deposit date: | 2015-05-04 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adhesive Dimerization of Human P-Cadherin Catalyzed by a Chaperone-like Mechanism
Structure, 24, 2016
|
|
4ZMQ
 
 | |
1GYR
 
 | | Mutant form of enoyl thioester reductase from Candida tropicalis | | Descriptor: | 2,4-DIENOYL-COA REDUCTASE, GLYCEROL, SULFATE ION | | Authors: | Airenne, T.T, Torkko, J.M, Van Der Plas, S, Sormunen, R.T, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2002-04-29 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
J.Mol.Biol., 327, 2003
|
|
2RK2
 
 | | DHFR R-67 complexed with NADP | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Dihydrofolate reductase type 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Krahn, J.M, London, R.E. | | Deposit date: | 2007-10-16 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a type II dihydrofolate reductase catalytic ternary complex.
Biochemistry, 46, 2007
|
|
2RK9
 
 | | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase superfamily member from Vibrio splendidus 12B01 | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Tyagi, R, Eswaramoorthy, S, Sauder, J.M, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-16 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase superfamily member from Vibrio splendidus 12B01.
To be Published
|
|
2X6B
 
 | | Potassium Channel from Magnetospirillum Magnetotacticum | | Descriptor: | ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL 10, BARIUM ION, PHOSPHOCHOLINE, ... | | Authors: | Clarke, O.B, Caputo, A.T, Hill, A.P, Vandenberg, J.I, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2010-02-15 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
8CYO
 
 | | Nurr1 Covalently Bound to a Synthetic Ligand, 10.25, via a Disulfide Bond | | Descriptor: | 2-(3,4-dichlorophenoxy)-N-(2-sulfanylethyl)acetamide, CHLORIDE ION, Nuclear receptor subfamily 4 group A member 2 | | Authors: | Bruning, J.M, Liu, J, England, P.M. | | Deposit date: | 2022-05-24 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Nurr1 Covalently Bound to a Synthetic Ligand, 10.25, via a Disulfide Bond
To Be Published
|
|
1H5W
 
 | | 2.1A Bacteriophage Phi-29 Connector | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, UPPER COLLAR PROTEIN | | Authors: | Guasch, A, Pous, J, Ibarra, B, Gomis-Ruth, F.X, Valpuesta, J.M, Sousa, N, Carrascosa, J.L, Coll, M. | | Deposit date: | 2001-05-28 | | Release date: | 2002-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detailed Architecture of a DNA Translocating Machine: The High-Resolution Structure of the Bacteriophage Phi29 Connector Particle.
J.Mol.Biol., 315, 2002
|
|
1YYL
 
 | | crystal structure of CD4M33, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 gp120 envelope glycoprotein and anti-HIV-1 antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD4M33, scorpion-toxin mimic of CD4, ... | | Authors: | Huang, C.C, Stricher, F, Martin, L, Decker, J.M, Majeed, S, Barthe, P, Hendrickson, W.A, Robinson, J, Roumestand, C, Sodroski, J, Wyatt, R, Shaw, G.M, Vita, C, Kwong, P.D. | | Deposit date: | 2005-02-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth.
Structure, 13, 2005
|
|
2X1C
 
 | | The crystal structure of precursor acyl coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum | | Descriptor: | ACYL-COENZYME, CHLORIDE ION, GLYCEROL, ... | | Authors: | Bokhove, M, Yoshida, H, Hensgens, C.M.H, van der Laan, J.M, Sutherland, J.D, Dijkstra, B.W. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of an Isopenicillin N Converting Ntn-Hydrolase Reveal Different Catalytic Roles for the Active Site Residues of Precursor and Mature Enzyme.
Structure, 18, 2010
|
|
1H2L
 
 | | Factor Inhibiting HIF-1 alpha in complex with HIF-1 alpha fragment peptide | | Descriptor: | 2-OXOGLUTARIC ACID, FACTOR INHIBITING HIF1, FE (II) ION, ... | | Authors: | Elkins, J.M, Hewitson, K.S, McNeill, L.A, Schlemminger, I, Seibel, J.F, Schofield, C.J. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor (Hif) Reveals Mechanism of Oxidative Modification of Hif-1Alpha
J.Biol.Chem., 278, 2003
|
|
2WYW
 
 | | High resolution structure of Thermus thermophilus enoyl-acyl carrier protein reductase NAD and triclosan-form | | Descriptor: | ENOYL-[ACYL CARRIER PROTEIN] REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Otero, J.M, Noel, A.J, Guardado-Calvo, P, Llamas-Saiz, A.L, van Raaij, M.J. | | Deposit date: | 2009-11-20 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structures of Thermus thermophilus enoyl-acyl carrier protein reductase in the apo form, in complex with NAD+ and in complex with NAD+ and triclosan.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 68, 2012
|
|
2WY8
 
 | | Staphylococcus aureus complement subversion protein Sbi-IV in complex with complement fragment C3d | | Descriptor: | COMPLEMENT C3D FRAGMENT, GLYCEROL, IGG-BINDING PROTEIN | | Authors: | Clark, E.A, Crennell, S, Upadhyay, A, Mackay, J.D, Bagby, S, van den Elsen, J.M. | | Deposit date: | 2009-11-13 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Structural Basis for Staphylococcal Complement Subversion: X-Ray Structure of the Complement- Binding Domain of Staphylococcus Aureus Protein Sbi in Complex with Ligand C3D.
Mol.Immunol., 48, 2011
|
|
2WSJ
 
 | | Crystal Structure of single point mutant Glu71Ser p-coumaric Acid Decarboxylase | | Descriptor: | BARIUM ION, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Rodriguez, H, Angulo, I, De Las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2009-09-07 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
4ZMT
 
 | |
1H1W
 
 | | High resolution crystal structure of the human PDK1 catalytic domain | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Biondi, R.M, Komander, D, Thomas, C.C, Lizcano, J.M, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2002-07-23 | | Release date: | 2003-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Crystal Structure of the Human Pdk1 Catalytic Domain Defines the Regulatory Phosphopeptide Docking Site
Embo J., 21, 2003
|
|
3ME7
 
 | | Crystal structure of putative electron transport protein aq_2194 from Aquifex aeolicus VF5 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative electron transport protein aq_2194 from Aquifex aeolicus VF5
To be Published
|
|
4ZN0
 
 | |
2X1E
 
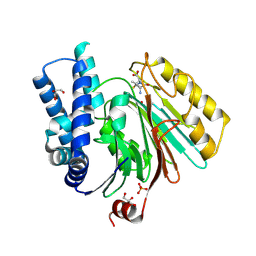 | | The crystal structure of mature acyl coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum in complex 6- aminopenicillanic acid | | Descriptor: | (2S,5R,6R)-6-AMINO-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, ACYL-COENZYME, CHLORIDE ION, ... | | Authors: | Bokhove, M, Yoshida, H, Hensgens, C.M.H, van der Laan, J.M, Sutherland, J.D, Dijkstra, B.W. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of an Isopenicillin N Converting Ntn-Hydrolase Reveal Different Catalytic Roles for the Active Site Residues of Precursor and Mature Enzyme.
Structure, 18, 2010
|
|
1GZ7
 
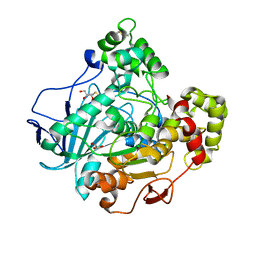 | | Crystal structure of the closed state of lipase 2 from Candida rugosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LIPASE 2 | | Authors: | Mancheno, J.M, Hermoso, J.A. | | Deposit date: | 2002-05-17 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Insights Into the Lipase/Esterase Behavior in the Candida Rugosa Lipases Family: Crystal Structure of the Lipase 2 Isoenzyme at 1.97A Resolution
J.Mol.Biol., 332, 2003
|
|
3K60
 
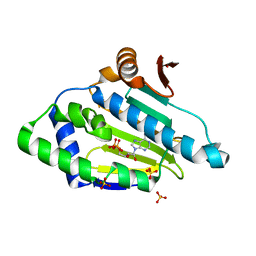 | |
3JBF
 
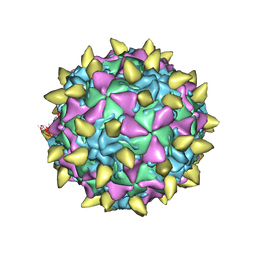 | | Complex of poliovirus with VHH PVSP19B | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
1HA6
 
 | | NMR Solution Structure of Murine CCL20/MIP-3a Chemokine | | Descriptor: | MACROPHAGE INFLAMMATORY PROTEIN 3 ALPHA | | Authors: | Perez-Canadillas, J.M, Zaballos, A, Gutierrez, J, Varona, R, Roncal, F, Albar, J.P, Marquez, G, Bruix, M. | | Deposit date: | 2001-03-28 | | Release date: | 2001-08-22 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Murine Ccl20/Mip3-A, a Chemiokine that Specifically Chemoattracs Immature Dendritic Cells and Lymphocytes Through its Highly Specific Interaction with the Beta-Chemokine Receptor Ccr6
J.Biol.Chem., 276, 2001
|
|
2BKS
 
 | | crystal structure of Renin-PF00074777 complex | | Descriptor: | (6R)-6-({[1-(3-HYDROXYPROPYL)-1,7-DIHYDROQUINOLIN-7-YL]OXY}METHYL)-1-(4-{3-[(2-METHOXYBENZYL)OXY]PROPOXY}PHENYL)PIPERAZIN-2-ONE, Renin | | Authors: | Powell, N.A, Clay, E.H, Holsworth, D.D, Edmunds, J.J, Bryant, J.W, Ryan, J.M, Jalaie, M, Zhang, E. | | Deposit date: | 2005-02-18 | | Release date: | 2006-04-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Equipotent Activity in Both Enantiomers of a Series of Ketopiperazine-Based Renin Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1PNN
 
 | | PEPTIDE NUCLEIC ACID (PNA) COMPLEXED WITH DNA | | Descriptor: | DNA (5'-D(GP*AP*AP*GP*AP*AP*GP*AP*G)-3'), PNA (NH2-P(*C*T*C*T*T*C*T*T*C-HIS-GLY-SER-SER-GLY-HIS-C*T*T*C*T*T*C*T*C)-COOH) | | Authors: | Betts, L, Veal, J.M. | | Deposit date: | 1995-10-13 | | Release date: | 1996-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Nucleic Acid Triple Helix Formed by a Peptide Nucleic Acid-DNA Complex
Science, 270, 1995
|
|
