2AB5
 
 | | bI3 LAGLIDADG Maturase | | Descriptor: | SULFATE ION, mRNA maturase | | Authors: | Longo, A, Leonard, C.W, Bassi, G.S, Berndt, D, Krahn, J.M, Hall, T.M, Weeks, K.M. | | Deposit date: | 2005-07-14 | | Release date: | 2005-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution from DNA to RNA recognition by the bI3 LAGLIDADG maturase
Nat.Struct.Mol.Biol., 12, 2005
|
|
3KML
 
 | |
1ZXQ
 
 | | THE CRYSTAL STRUCTURE OF ICAM-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, INTERCELLULAR ADHESION MOLECULE-2 | | Authors: | Casasnovas, J.M, Springer, T.A, Harrison, S.C, Wang, J.-H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of ICAM-2 reveals a distinctive integrin recognition surface.
Nature, 387, 1997
|
|
1ICA
 
 | |
1ZZY
 
 | |
2Q7H
 
 | | Pyrrolysyl-tRNA synthetase bound to adenylated pyrrolysine and pyrophosphate | | Descriptor: | (2R)-2-AMINO-6-({[(2S,3R)-3-METHYLPYRROLIDIN-2-YL]CARBONYL}AMINO)HEXANOYL [(2S,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL HYDROGEN (R)-PHOSPHATE, 1,2-ETHANEDIOL, PYROPHOSPHATE 2-, ... | | Authors: | Kavran, J.M, Steitz, T.A. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1I1J
 
 | | STRUCTURE OF MELANOMA INHIBITORY ACTIVITY PROTEIN: A MEMBER OF A NEW FAMILY OF SECRETED PROTEINS | | Descriptor: | MELANOMA DERIVED GROWTH REGULATORY PROTEIN | | Authors: | Lougheed, J.C, Holton, J.M, Alber, T, Bazan, J.F, Handel, T.M. | | Deposit date: | 2001-02-02 | | Release date: | 2001-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structure of melanoma inhibitory activity protein, a member of a recently identified family of secreted proteins.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2PFO
 
 | | DNA Polymerase lambda in complex with DNA and dUPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA polymerase lambda, ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2007-04-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of the catalytic metal during polymerization by DNA polymerase lambda.
DNA Repair, 6, 2007
|
|
2A11
 
 | |
4XG1
 
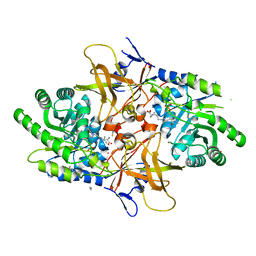 | | Psychromonas ingrahamii diaminopimelate decarboxylase with LLP | | Descriptor: | (2S)-2-amino-6-[[3-hydroxy-2-methyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]hexanoic acid, Diaminopimelate decarboxylase, POTASSIUM ION, ... | | Authors: | Peverelli, M.G, Wubben, J.M, Panjikar, S, Perugini, M.A. | | Deposit date: | 2014-12-30 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Expression to crystallization of diaminopimelate decarboxylase from the psychrophile Psychromonas ingrahamii
To Be Published
|
|
3L49
 
 | | CRYSTAL STRUCTURE OF ABC SUGAR TRANSPORTER SUBUNIT FROM Rhodobacter sphaeroides 2.4.1 | | Descriptor: | ABC sugar (Ribose) transporter, periplasmic substrate-binding subunit, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Ozyurt, S, Dickey, M, Do, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-05 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF ABC SUGAR TRANSPORTER FROM Rhodobacter sphaeroides
To be Published
|
|
2Q08
 
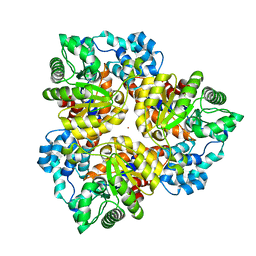 | | Crystal structure of the protein BH0493 from Bacillus halodurans C-125 complexed with ZN | | Descriptor: | BH0493 protein, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Patskovsky, Y, Sauder, J.M, Burley, S.K, Raushel, F, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-21 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Glucoronate Isomerase from Bacillus halodurans.
To be Published
|
|
2YIB
 
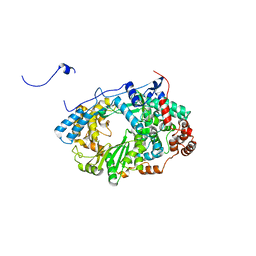 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus | | Descriptor: | RNA-DIRECTED RNA POLYMERASE | | Authors: | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
3KPT
 
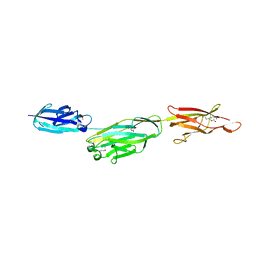 | | Crystal structure of BcpA, the major pilin subunit of Bacillus cereus | | Descriptor: | CALCIUM ION, Collagen adhesion protein | | Authors: | Poor, C.B, Budzik, J.M, Schneewind, O, He, C. | | Deposit date: | 2009-11-16 | | Release date: | 2009-11-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Intramolecular amide bonds stabilize pili on the surface of bacilli.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1GQW
 
 | | Taurine/alpha-ketoglutarate Dioxygenase from Escherichia coli | | Descriptor: | 2-AMINOETHANESULFONIC ACID, 2-OXOGLUTARIC ACID, ALPHA-KETOGLUTARATE-DEPENDENT TAURINE DIOXYGENASE, ... | | Authors: | Elkins, J.M, Ryle, M.J, Clifton, I.J, Dunning-Hotopp, J.C, Lloyd, J.S, Burzlaff, N.I, Baldwin, J.E, Hausinger, R.P, Roach, P.L. | | Deposit date: | 2001-12-05 | | Release date: | 2002-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Crystal Structure of Escherichia Coli Taurine/Alpha-Ketoglutarate Dioxygenase Complexed to Ferrous Iron and Substrates
Biochemistry, 41, 2002
|
|
2YME
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with granisetron | | Descriptor: | 1-methyl-N-[(1R,5S)-9-methyl-9-azabicyclo[3.3.1]nonan-3-yl]indazole-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht3 Receptors.
Embo Rep., 14, 2013
|
|
2AAN
 
 | |
2PFJ
 
 | | Crystal Structure of T7 Endo I resolvase in complex with a Holliday Junction | | Descriptor: | 27-MER, CALCIUM ION, Endodeoxyribonuclease 1 | | Authors: | Hadden, J.M, Declais, A.C, Carr, S.B, Lilley, D.M, Phillips, S.E. | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis of Holliday junction resolution by T7 endonuclease I.
Nature, 449, 2007
|
|
2PX5
 
 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH (Orthorhombic crystal form) | | Descriptor: | Genome polyprotein [Contains: Capsid protein C (Core protein); Envelope protein M (Matrix protein); Major envelope protein E; Non-structural protein 1 (NS1); Non-structural protein 2A (NS2A); Flavivirin protease NS2B regulatory subunit; Flavivirin protease NS3 catalytic subunit; Non-structural protein 4A (NS4A); Non-structural protein 4B (NS4B); RNA-directed RNA polymerase (EC 2.7.7.48) (NS5)], S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
3L4J
 
 | | Topoisomerase II-DNA cleavage complex, apo | | Descriptor: | 3'-THIO-THYMIDINE-5'-PHOSPHATE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*CP*GP*TP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*TP*AP*GP*CP*AP*GP*TP*AP*GP*G)-3'), ... | | Authors: | Schmidt, B.H, Burgin, A.B, Deweese, J.E, Osheroff, N, Berger, J.M. | | Deposit date: | 2009-12-20 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases.
Nature, 465, 2010
|
|
3KPD
 
 | | Crystal Structure of the CBS domain pair of protein MJ0100 in complex with 5 -methylthioadenosine and S-adenosyl-L-methionine. | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, S-ADENOSYLMETHIONINE, Uncharacterized protein MJ0100 | | Authors: | Lucas, M, Oyenarte, I, Garcia, I.G, Arribas, E.A, Encinar, J.A, Kortazar, D, Fernandez, J.A, Mato, J.M, Martinez-Chantar, M.L, Martinez-Cruz, L.A. | | Deposit date: | 2009-11-16 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Binding of S-Methyl-5'-Thioadenosine and S-Adenosyl-l-Methionine to Protein MJ0100 Triggers an Open-to-Closed Conformational Change in Its CBS Motif Pair.
J.Mol.Biol., 396, 2010
|
|
3KL2
 
 | | Crystal structure of a putative isochorismatase from Streptomyces avermitilis | | Descriptor: | Putative isochorismatase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-06 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative isochorismatase from Streptomyces avermitilis
To be Published
|
|
2WKS
 
 | | Structure of Helicobacter pylori Type II Dehydroquinase with a new carbasugar-thiophene inhibitor. | | Descriptor: | (1R,4S,5R)-1,4,5-trihydroxy-3-[(5-methyl-1-benzothiophen-2-yl)methoxy]cyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Otero, J.M, Guardado-Calvo, P, Llamas-Saiz, A.L, Prazeres, V.F.V, Tizon, L, Castedo, L, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2009-06-17 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synthesis and biological evaluation of new nanomolar competitive inhibitors of Helicobacter pylori type II dehydroquinase. Structural details of the role of the aromatic moieties with essential residues.
J. Med. Chem., 53, 2010
|
|
2X79
 
 | | Inward facing conformation of Mhp1 | | Descriptor: | Hydantoin permease | | Authors: | Shimamura, T, Weyand, S, Beckstein, O, Rutherford, N.G, Hadden, J.M, Sharples, D, Sansom, M.S.P, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Basis of Alternating Access Membrane Transport by the Sodium-Hydantoin Transporter Mhp1.
Science, 328, 2010
|
|
2WLY
 
 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitotrio-thiazoline. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-29 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chitinase inhibition by chitobiose and chitotriose thiazolines.
Angew. Chem. Int. Ed. Engl., 49, 2010
|
|
