5I40
 
 | | BRD9 in complex with Cpd1 (6-methyl-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one) | | Descriptor: | 1,2-ETHANEDIOL, 6-methyl-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one, Bromodomain-containing protein 9, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.0402 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
4NFO
 
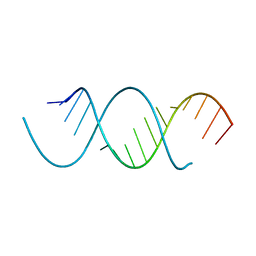 | | Crystal Structure Analysis of the 16mer GCAGACUUAAGUCUGC | | Descriptor: | GCAGACUUAAGUCUGC, SPERMINE | | Authors: | Beal, P.A, Fisher, A.J, Phelps, K.J, Ibarra-Soza, J.M, Zheng, Y. | | Deposit date: | 2013-10-31 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Click Modification of RNA at Adenosine: Structure and Reactivity of 7-Ethynyl- and 7-Triazolyl-8-aza-7-deazaadenosine in RNA.
Acs Chem.Biol., 9, 2014
|
|
4NDB
 
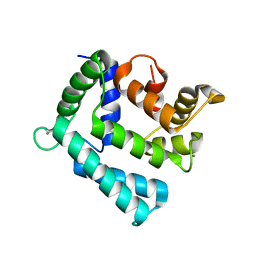 | | X-ray structure of a mutant (T61D) of calexcitin - a neuronal calcium-signalling protein | | Descriptor: | CALCIUM ION, Calexcitin | | Authors: | Erskine, P.T, Fokas, A, Muriithi, C, Razzall, E, Bowyer, A, Findlow, I.S, Werner, J.M, Wallace, B.A, Wood, S.P, Cooper, J.B. | | Deposit date: | 2013-10-25 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray, spectroscopic and normal-mode dynamics of calexcitin: structure-function studies of a neuronal calcium-signalling protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4P37
 
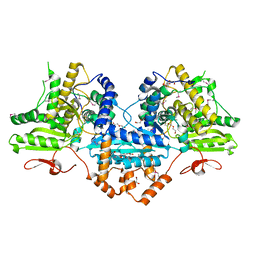 | | Crystal structure of the Megavirus polyadenylate synthase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Priet, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2014-03-06 | | Release date: | 2015-04-01 | | Last modified: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | mRNA maturation in giant viruses: variation on a theme.
Nucleic Acids Res., 43, 2015
|
|
2X7L
 
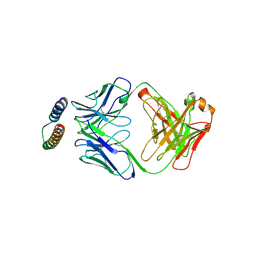 | | Implications of the HIV-1 Rev dimer structure at 3.2A resolution for multimeric binding to the Rev response element | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, PROTEIN REV | | Authors: | DiMattia, M.A, Watts, N.R, Stahl, S.J, Rader, C, Wingfield, P.T, Stuart, D.I, Steven, A.C, Grimes, J.M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Implications of the HIV-1 Rev Dimer Structure at 3. 2 A Resolution for Multimeric Binding to the Rev Response Element.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4P5Y
 
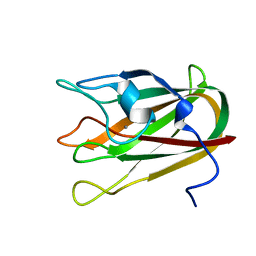 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with N-acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, Glycosyl hydrolase, ... | | Authors: | Grondin, J.M, Allingham, J.S, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-03-04 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
PLoS ONE, 12, 2017
|
|
4PFM
 
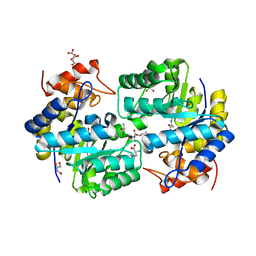 | | SHEWANELLA BENTHICA DHDPS WITH LYSINE AND PYRUVATE | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CITRATE ANION, GLYCEROL, ... | | Authors: | Wubben, J.M, Paxman, J.J, Dogovski, C, Panjikar, S, Perugini, M.A. | | Deposit date: | 2014-04-30 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Defining a Molecular Determinant of Lysine-Mediated Allosteric Inhibition of DHDPS.
To Be Published
|
|
6T9U
 
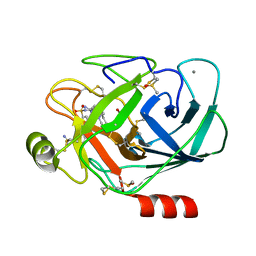 | | Bovine Trypsine in complex with the synthetic inhibitor (S)-3'-(N-(1-(4-(3-(tert-butyl)ureido)piperidin-1-yl)-3-(3-carbamimidoylphenyl)-1-oxopropan-2-yl)sulfamoyl)-[1,1'-biphenyl]-3-carboximidamide (MI-490) | | Descriptor: | 1-~{tert}-butyl-3-[1-[(2~{S})-3-(3-carbamimidoylphenyl)-2-[[3-(3-carbamimidoylphenyl)phenyl]sulfonylamino]propanoyl]piperidin-4-yl]urea, CALCIUM ION, Cationic Trypsin, ... | | Authors: | Mueller, J.M, Merkl, S, Keils, A, Pilgram, O, Steinmetzer, T. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.067509 Å) | | Cite: | Improving the selectivity of 3-amidinophenylalanine-derived matriptase inhibitors
Eur.J.Med.Chem., 2022
|
|
2W2S
 
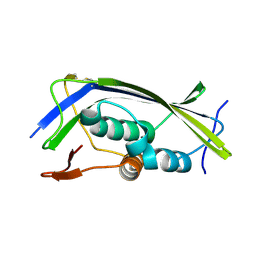 | | Structure of the Lagos bat virus matrix protein | | Descriptor: | MATRIX PROTEIN | | Authors: | Graham, S.C, Assenberg, R, Delmas, O, Verma, A, Gholami, A, Talbi, C, Owens, R.J, Stuart, D.I, Grimes, J.M, Bourhy, H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rhabdovirus Matrix Protein Structures Reveal a Novel Mode of Self-Association.
Plos Pathog., 4, 2008
|
|
4MAF
 
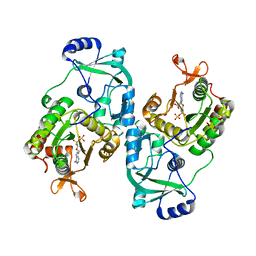 | | Soybean ATP Sulfurylase | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, ATP sulfurylase | | Authors: | Herrmann, J, Ravilious, G.E, McKinney, S.E, Westfall, C.S, Lee, S.G, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2013-08-16 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and mechanism of soybean ATP sulfurylase and the committed step in plant sulfur assimilation.
J.Biol.Chem., 289, 2014
|
|
6T9V
 
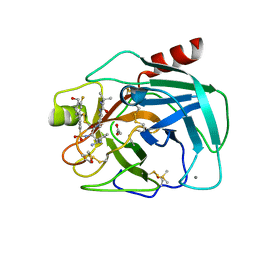 | | Bovine Trypsin in complex with the synthetic inhibitor (S)-3-(3-(4-(3-(tert-butyl)ureido)piperidin-1-yl)-2-((3'-fluoro-4'-(hydroxymethyl)-[1,1'-biphenyl])-3-sulfonamido)-3-oxopropyl)benzimidamide (MI-1904) | | Descriptor: | 1-~{tert}-butyl-3-[1-[(2~{S})-3-(3-carbamimidoylphenyl)-2-[[3-[3-fluoranyl-4-(hydroxymethyl)phenyl]phenyl]sulfonylamino ]propanoyl]piperidin-4-yl]urea, CALCIUM ION, Cationic Trypsin, ... | | Authors: | Merkl, S, Keils, A, Mueller, J.M, Pilgram, O, Steinmetzer, T. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12642729 Å) | | Cite: | Improving the selectivity of 3-amidinophenylalanine-derived matriptase inhibitors
Eur.J.Med.Chem., 2022
|
|
3CCP
 
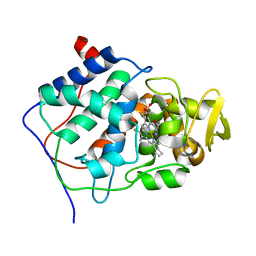 | | X-RAY STRUCTURES OF RECOMBINANT YEAST CYTOCHROME C PEROXIDASE AND THREE HEME-CLEFT MUTANTS PREPARED BY SITE-DIRECTED MUTAGENESIS | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, YEAST CYTOCHROME C PEROXIDASE | | Authors: | Wang, J, Mauro, J.M, Edwards, S.L, Oatley, S.J, Fishel, L.A, Ashford, V.A, Xuong, N.-H, Kraut, J. | | Deposit date: | 1990-02-28 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of recombinant yeast cytochrome c peroxidase and three heme-cleft mutants prepared by site-directed mutagenesis.
Biochemistry, 29, 1990
|
|
4LXA
 
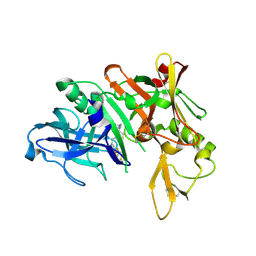 | | Crystal Structure of Human Beta Secretase in Complex with Compound 11a | | Descriptor: | (1R,3S,4S,5R)-3-{4-amino-3-fluoro-5-[(1,1,1,3,3,3-hexafluoropropan-2-yl)oxy]benzyl}-5-[(3-tert-butylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1-oxide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2013-07-29 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of cyclic sulfoxide hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4LXV
 
 | | Crystal Structure of the Hemagglutinin from a H1N1pdm A/WASHINGTON/5/2011 virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Chang, J.C, Guo, Z, Carney, P.J, Shore, D.A, Donis, R.O, Cox, N.J, Villanueva, J.M, Klimov, A.I, Stevens, J. | | Deposit date: | 2013-07-30 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Stability of Influenza A(H1N1)pdm09 Virus Hemagglutinins.
J.Virol., 88, 2014
|
|
4M0A
 
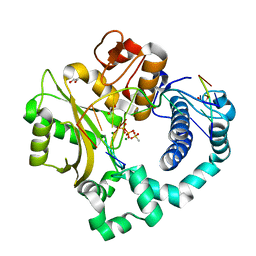 | | Human DNA Polymerase Mu post-catalytic complex | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA-directed DNA/RNA polymerase mu, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2013-08-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Sustained active site rigidity during synthesis by human DNA polymerase mu.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1WB0
 
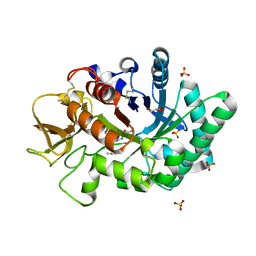 | | specificity and affinity of natural product cyclopentapeptide inhibitor Argifin against human chitinase | | Descriptor: | ARGIFIN, CHITOTRIOSIDASE 1, GLYCEROL, ... | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Hodkinson, M, Adams, D.J, Shiomi, K, Omura, S, Van Aalten, D.M.F. | | Deposit date: | 2004-10-29 | | Release date: | 2005-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Specificity and Affinity of Natural Product Cyclopentapeptide Inhibitors Against Aspergillus Fumigatus, Human and Bacterial Chitinases
Chem.Biol., 12, 2005
|
|
4M21
 
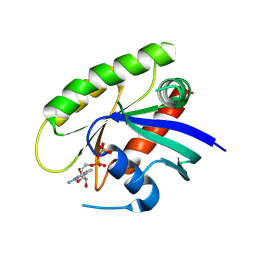 | | Crystal Structure of small molecule acrylamide 11 covalently bound to K-Ras G12C | | Descriptor: | 1-(4-{[(4,5-dichloro-2-methoxyphenyl)amino]acetyl}piperazin-1-yl)propan-1-one, GUANOSINE-5'-DIPHOSPHATE, K-Ras GTPase | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4LYF
 
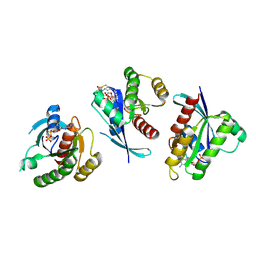 | | Crystal Structure of small molecule vinylsulfonamide 8 covalently bound to K-Ras G12C | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, N-{1-[N-(4,5-dichloro-2-hydroxyphenyl)glycyl]piperidin-4-yl}ethanesulfonamide | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-31 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.568 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
2JVG
 
 | | Structure of C3-binding domain 4 of Staphylococcus aureus protein Sbi | | Descriptor: | IgG-binding protein SBI | | Authors: | Upadhyay, A, Burman, J, Clark, E.A, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi.
J.Biol.Chem., 283, 2008
|
|
3JCL
 
 | | Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer | | Descriptor: | Spike glycoprotein | | Authors: | Walls, A.C, Tortorici, M.A, Bosch, B.J, Frenz, B, Rottier, P.J.M, DiMaio, F, Rey, F.A, Veesler, D. | | Deposit date: | 2015-12-21 | | Release date: | 2016-02-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer.
Nature, 531, 2016
|
|
4PQI
 
 | | Crystal structure of glutathione transferase lambda3 from Populus trichocarpa | | Descriptor: | CALCIUM ION, GLUTATHIONE, In2-1 family protein, ... | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Prosper, P, Didierjean, C, Haouz, A, Saul, F, Rouhier, N, Hecker, A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and enzymatic insights into Lambda glutathione transferases from Populus trichocarpa, monomeric enzymes constituting an early divergent class specific to terrestrial plants.
Biochem.J., 462, 2014
|
|
2JVH
 
 | | Structure of C3-binding domain 4 of S. aureus protein Sbi | | Descriptor: | IgG-binding protein SBI | | Authors: | Upadhyay, A, Burman, J, Clark, E.A, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi.
J.Biol.Chem., 283, 2008
|
|
1LAX
 
 | | CRYSTAL STRUCTURE OF MALE31, A DEFECTIVE FOLDING MUTANT OF MALTOSE-BINDING PROTEIN | | Descriptor: | MALTOSE-BINDING PROTEIN MUTANT MALE31, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Saul, F.A, Mourez, M, Vulliez-le Normand, B, Sassoon, N, Bentley, G.A, Betton, J.M. | | Deposit date: | 2002-03-29 | | Release date: | 2003-03-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a defective folding protein
PROTEIN SCI., 12, 2003
|
|
4MX1
 
 | | Structure of ricin A chain bound with 2-amino-4-oxo-N-(2-(3-phenylureido)ethyl)-3,4-dihydropteridine-7-carboxamide | | Descriptor: | 2-amino-4-oxo-N-{2-[(phenylcarbamoyl)amino]ethyl}-3,4-dihydropteridine-7-carboxamide, MALONIC ACID, Ricin A chain, ... | | Authors: | Robertus, J.D, Wiget, P.A, Manzano, L.A, Pruet, J.M, Gao, G, Saito, R, Jasheway, K.R, Monzingo, A.F, Anslyn, E.V. | | Deposit date: | 2013-09-25 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Sulfur incorporation generally improves Ricin inhibition in pterin-appended glycine-phenylalanine dipeptide mimics.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4N4A
 
 | | Cystal structure of Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 | | Authors: | Smietanski, M, Werener, M, Purta, E, Kaminska, K.H, Stepinski, J, Darzynkiewicz, E, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.
Nat Commun, 5, 2014
|
|
