6B8B
 
 | | E. coli LptB in complex with ADP and a novobiocin derivative | | Descriptor: | (3s,5s,7s)-N-{7-[(3-O-carbamoyl-6-deoxy-5-methyl-4-O-methyl-beta-D-gulopyranosyl)oxy]-4-hydroxy-8-methyl-2-oxo-2H-1-ben zopyran-3-yl}tricyclo[3.3.1.1~3,7~]decane-1-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, ... | | Authors: | Mandler, M.D, Owens, T.W, Lazarus, M.B, May, J.M, Kahne, D.K. | | Deposit date: | 2017-10-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Antibiotic Novobiocin Binds and Activates the ATPase That Powers Lipopolysaccharide Transport.
J. Am. Chem. Soc., 139, 2017
|
|
3BCU
 
 | | Glucogen Phosphorylase complex with thymidine | | Descriptor: | Glycogen phosphorylase, muscle form, THYMIDINE | | Authors: | Sovantzis, D.A, Hadjiloi, T, Hayes, J.M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G. | | Deposit date: | 2007-11-13 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | D-Glucopyranosyl pyrimidine nucleoside binding to muscle glycogen phosphorylase b
To be Published
|
|
8E1K
 
 | |
8DZX
 
 | |
3B1Y
 
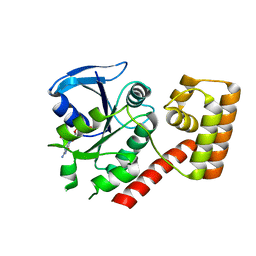 | | Crystal structure of an S. thermophilus NFeoB T35A mutant bound to GDP | | Descriptor: | AMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ferrous iron uptake transporter protein B | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-07-15 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A suite of Switch I and Switch II mutant structures from the G-protein domain of FeoB
Acta Crystallogr.,Sect.D, 67, 2011
|
|
8E09
 
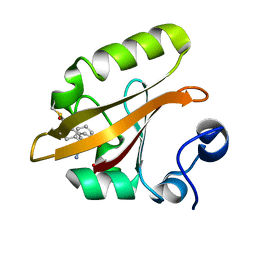 | |
8DZY
 
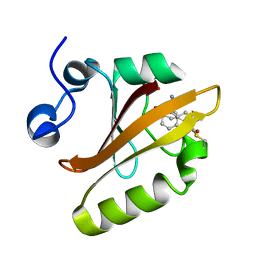 | |
8E02
 
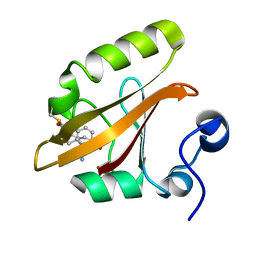 | |
8E1L
 
 | |
8E03
 
 | |
8DZU
 
 | |
3BIA
 
 | | Tim-4 in complex with sodium potassium tartrate | | Descriptor: | L(+)-TARTARIC ACID, SODIUM ION, T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Freeman, G.J, Casasnovas, J.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Protein 4 Show a Metal-Ion-Dependent Ligand Binding Site where Phosphatidylserine Binds.
Immunity, 27, 2007
|
|
6AUD
 
 | | PI3K-gamma K802T in complex with Cpd 8 10-((1-(tert-butyl)piperidin-4-yl)sulfinyl)-2-(1-isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepine | | Descriptor: | 10-[(S)-(1-tert-butylpiperidin-4-yl)sulfinyl]-2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Ultsch, M. | | Deposit date: | 2017-08-31 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Design of Selective Benzoxazepin PI3K delta Inhibitors Through Control of Dihedral Angles.
ACS Med Chem Lett, 8, 2017
|
|
3BDA
 
 | | Glycogen Phosphorylase complex with 1(-D-glucopyranosyl) cyanuric acid | | Descriptor: | 1-beta-D-glucopyranosyl-1,3,5-triazinane-2,4,6-trione, Glycogen phosphorylase, muscle form, ... | | Authors: | Sovantzis, D.A, Hadjiloi, T, Hayes, J.M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D-Glucopyranosyl pyrimidine nucleoside binding to muscle glycogen phosphorylase b
To be Published
|
|
3BDY
 
 | | Dual specific bH1 Fab in complex with VEGF | | Descriptor: | Fab Fragment -Heavy Chain, Fab Fragment -Light Chain, GLYCEROL, ... | | Authors: | Bostrom, J.M, Wiesmann, C, Appleton, B.A. | | Deposit date: | 2007-11-15 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Variants of the antibody herceptin that interact with HER2 and VEGF at the antigen binding site
Science, 323, 2009
|
|
6BE0
 
 | | AvrA delL154 with IP6, CoA | | Descriptor: | AvrA, COENZYME A, INOSITOL HEXAKISPHOSPHATE | | Authors: | Labriola, J.M, Nagar, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural Analysis of the Bacterial Effector AvrA Identifies a Critical Helix Involved in Substrate Recognition.
Biochemistry, 57, 2018
|
|
3BD6
 
 | | Glycogen Phosphorylase complex with 1(-D-ribofuranosyl) cyanuric acid | | Descriptor: | 1-beta-D-ribofuranosyl-1,3,5-triazinane-2,4,6-trione, Glycogen phosphorylase, muscle form | | Authors: | Sovantzis, D.A, Hadjiloi, T, Hayes, J.M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D-Glucopyranosyl pyrimidine nucleoside binding to muscle glycogen phosphorylase b
To be Published
|
|
8E9L
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9N
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIY in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9M
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT bound to maleic acid at 278 K | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9R
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFCS in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9K
 
 | | Crystal structure of wild-type E. coli aspartate aminotransferase bound to maleate at 278 K | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9Q
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX bound to maleic acid at 278 K | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9O
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIY bound to maleic acid at 278 K | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
3B2N
 
 | | Crystal structure of DNA-binding response regulator, LuxR family, from Staphylococcus aureus | | Descriptor: | SODIUM ION, Uncharacterized protein Q99UF4 | | Authors: | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-18 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of DNA-binding response regulator, LuxR family, from Staphylococcus aureus.
To be Published
|
|
