2ODE
 
 | | Crystal structure of the heterodimeric complex of human RGS8 and activated Gi alpha 3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(k) subunit alpha, MAGNESIUM ION, ... | | Authors: | Gileadi, C, Soundararajan, M, Turnbull, A.P, Elkins, J.M, Papagrigoriou, E, Pike, A.C.W, Bunkoczi, G, Gorrec, F, Umeano, C, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C.H, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-22 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4OFS
 
 | | Crystal structure of a truncated catalytic core of the 2-oxoacid dehydrogenase multienzyme complex from Thermoplasma acidophilum | | Descriptor: | Probable lipoamide acyltransferase | | Authors: | Marrot, N.L, Marshall, J.J.T, Svergun, D.I, Crennell, S.J, Hough, D.W, van den Elsen, J.M.H, Danson, M.J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Why are the 2-oxoacid dehydrogenase complexes so large? Generation of an active trimeric complex.
Biochem.J., 463, 2014
|
|
4P2G
 
 | | Crystal structure of DJ-1 in sulfinic acid form (aged crystal) | | Descriptor: | 1,2-ETHANEDIOL, PENTAETHYLENE GLYCOL, Protein DJ-1 | | Authors: | Tashiro, S, Wu, C.-X, Hoang, Q.Q, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Thermodynamic and Structural Characterization of the Specific Binding of Zn(II) to Human Protein DJ-1.
Biochemistry, 53, 2014
|
|
4P36
 
 | | Crystal structure of DJ-1 With Zn(II) bound (crystal 2) | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, Protein DJ-1, ZINC ION | | Authors: | Tashiro, S, Wu, C.-X, Hoang, Q.Q, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.182 Å) | | Cite: | Thermodynamic and Structural Characterization of the Specific Binding of Zn(II) to Human Protein DJ-1.
Biochemistry, 53, 2014
|
|
4P35
 
 | | Crystal structure of DJ-1 with Zinc(II) bound (crystal I) | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein DJ-1, ... | | Authors: | Tashiro, S, Wu, C.-X, Hoang, Q.Q, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thermodynamic and Structural Characterization of the Specific Binding of Zn(II) to Human Protein DJ-1.
Biochemistry, 53, 2014
|
|
1BIO
 
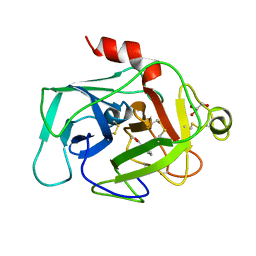 | | HUMAN COMPLEMENT FACTOR D IN COMPLEX WITH ISATOIC ANHYDRIDE INHIBITOR | | Descriptor: | COMPLEMENT FACTOR D, GLYCEROL, ISATOIC ANHYDRIDE | | Authors: | Jing, H, Babu, Y.S, Moore, D, Kilpatrick, J.M, Liu, X.-Y, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity.
J.Mol.Biol., 282, 1998
|
|
3RDC
 
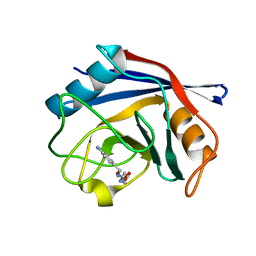 | | Human Cyclophilin D Complexed with an Inhibitor | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ethyl N-[(4-aminobenzyl)carbamoyl]glycinate | | Authors: | Colliandre, L, Ahmed-Belkacem, H, Bessin, Y, Pawlotsky, J.M, Guichou, J.F. | | Deposit date: | 2011-04-01 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Combining 'dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1AL2
 
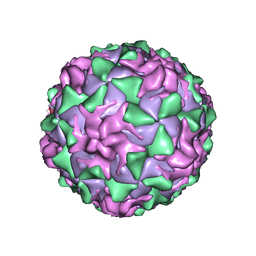 | | P1/MAHONEY POLIOVIRUS, SINGLE SITE MUTANT V1160I | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-06-09 | | Release date: | 1997-11-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AMP
 
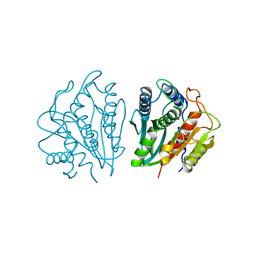 | | CRYSTAL STRUCTURE OF AEROMONAS PROTEOLYTICA AMINOPEPTIDASE: A PROTOTYPICAL MEMBER OF THE CO-CATALYTIC ZINC ENZYME FAMILY | | Descriptor: | AMINOPEPTIDASE, ZINC ION | | Authors: | Chevrier, B, Schalk, C, D'Orchymont, H, Rondeau, J.M, Moras, D, Tarnus, C. | | Deposit date: | 1994-04-22 | | Release date: | 1994-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Aeromonas proteolytica aminopeptidase: a prototypical member of the co-catalytic zinc enzyme family.
Structure, 2, 1994
|
|
1BJT
 
 | | TOPOISOMERASE II RESIDUES 409-1201 | | Descriptor: | TOPOISOMERASE II | | Authors: | Fass, D, Bogden, C.E, Berger, J.M. | | Deposit date: | 1998-06-29 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands.
Nat.Struct.Biol., 6, 1999
|
|
1B65
 
 | | Structure of l-aminopeptidase d-ala-esterase/amidase from ochrobactrum anthropi, a prototype for the serine aminopeptidases, reveals a new variant among the ntn hydrolase fold | | Descriptor: | PROTEIN (AMINOPEPTIDASE) | | Authors: | Bompard-Gilles, C, Villeret, V, Davies, G.J, Fanuel, L, Joris, B, Frere, J.M, Van Beeumen, J. | | Deposit date: | 1999-01-20 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A new variant of the Ntn hydrolase fold revealed by the crystal structure
of L-aminopeptidase D-ala-esterase/amidase from Ochrobactrum anthropi.
Structure Fold.Des., 8, 2000
|
|
1AR6
 
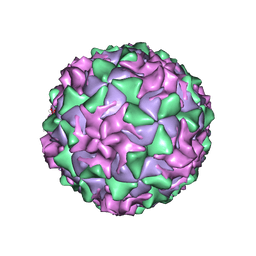 | | P1/MAHONEY POLIOVIRUS, DOUBLE MUTANT V1160I +P1095S | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1BL8
 
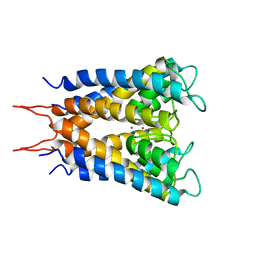 | | POTASSIUM CHANNEL (KCSA) FROM STREPTOMYCES LIVIDANS | | Descriptor: | POTASSIUM ION, PROTEIN (POTASSIUM CHANNEL PROTEIN) | | Authors: | Doyle, D.A, Cabral, J.M, Pfuetzner, R.A, Kuo, A, Gulbis, J.M, Cohen, S.L, Chait, B.T, Mackinnon, R. | | Deposit date: | 1998-07-23 | | Release date: | 1998-07-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the potassium channel: molecular basis of K+ conduction and selectivity.
Science, 280, 1998
|
|
4P34
 
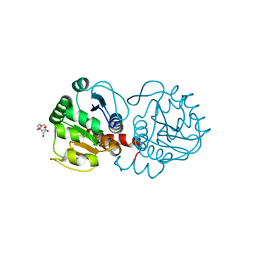 | | Crystal structure of DJ-1 in sulfenic acid form (fresh crystal) | | Descriptor: | PENTAETHYLENE GLYCOL, Protein DJ-1 | | Authors: | Tashiro, S, Wu, C.-X, Hoang, Q.Q, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Thermodynamic and Structural Characterization of the Specific Binding of Zn(II) to Human Protein DJ-1.
Biochemistry, 53, 2014
|
|
1AR9
 
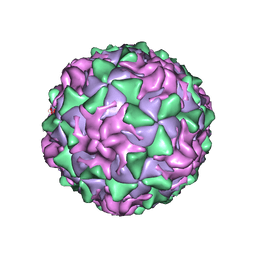 | | P1/MAHONEY POLIOVIRUS, SINGLE SITE MUTANT H2142Y | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
3ZX8
 
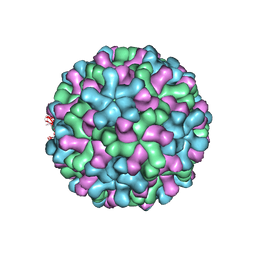 | | Cryo-EM reconstruction of native and expanded Turnip Crinkle virus | | Descriptor: | CAPSID PROTEIN | | Authors: | Bakker, S.E, Robottom, J, Hogle, J.M, Maeda, A, Pearson, A.R, Stockley, P.G, Ranson, N.A, Harrison, S.C. | | Deposit date: | 2011-08-08 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Isolation of an Asymmetric RNA Uncoating Intermediate for a Single-Stranded RNA Plant Virus.
J.Mol.Biol., 417, 2012
|
|
1C54
 
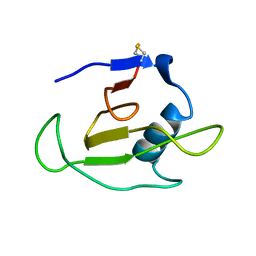 | | SOLUTION STRUCTURE OF RIBONUCLEASE SA | | Descriptor: | RIBONUCLEASE SA | | Authors: | Laurents, D.V, Canadillas-Perez, J.M, Santoro, J, Schell, D, Pace, C.N, Rico, M, Bruix, M. | | Deposit date: | 1999-10-22 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ribonuclease Sa.
Proteins, 44, 2001
|
|
1CCP
 
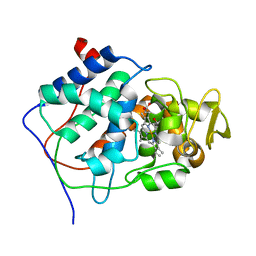 | | X-RAY STRUCTURES OF RECOMBINANT YEAST CYTOCHROME C PEROXIDASE AND THREE HEME-CLEFT MUTANTS PREPARED BY SITE-DIRECTED MUTAGENESIS | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, YEAST CYTOCHROME C PEROXIDASE | | Authors: | Wang, J, Mauro, J.M, Edwards, S.L, Oatley, S.J, Fishel, L.A, Ashford, V.A, Xuong, N.-H, Kraut, J. | | Deposit date: | 1990-02-28 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of recombinant yeast cytochrome c peroxidase and three heme-cleft mutants prepared by site-directed mutagenesis.
Biochemistry, 29, 1990
|
|
1AR7
 
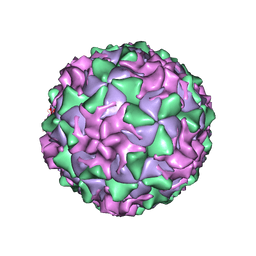 | | P1/MAHONEY POLIOVIRUS, DOUBLE MUTANT P1095S + H2142Y | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AR8
 
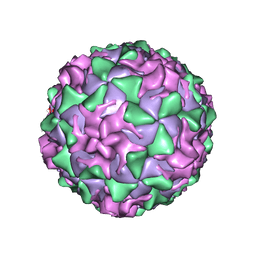 | | P1/MAHONEY POLIOVIRUS, MUTANT P1095S | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1ASJ
 
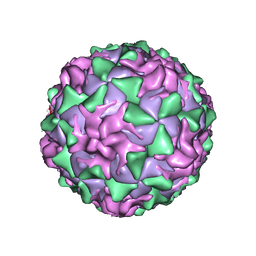 | | P1/MAHONEY POLIOVIRUS, AT CRYOGENIC TEMPERATURE | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
4Q97
 
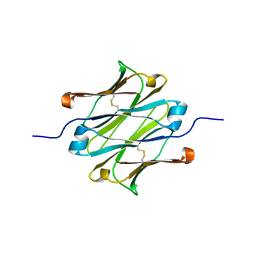 | | IgNAR antibody domain C1 | | Descriptor: | Novel antigen receptor | | Authors: | Feige, J.M, Graewert, M.A, Marcinowski, M, Hennig, J, Behnke, J, Auslaender, D, Herold, E.M, Peschek, J, Castro, C.D, Flajnik, M.F, Hendershot, L.M, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2014-04-29 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3F8C
 
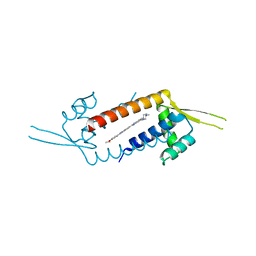 | | Crystal structure of multidrug binding transcriptional regulator LmrR complexed with Hoechst 33342 | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, Transcriptional regulator, PadR-like family | | Authors: | Madoori, P.K, Agustiandari, H, Driessen, A.J.M, Thunnissen, A.-M.W.H. | | Deposit date: | 2008-11-12 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition.
Embo J., 28, 2009
|
|
3F8B
 
 | | Crystal structure of the multidrug binding transcriptional regulator LmrR in drug free state | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Madoori, P.K, Agustiandari, H, Driessen, A.J.M, Thunnissen, A.-M.W.H. | | Deposit date: | 2008-11-12 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition.
Embo J., 28, 2009
|
|
1OUW
 
 | | Crystal structure of Calystegia sepium agglutinin | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, IMIDAZOLE, ... | | Authors: | Bourne, Y, Roig-Zamboni, V, Barre, A, Peumans, W.J, Astoul, C.H, van Damme, E.J.M, Rouge, P. | | Deposit date: | 2003-03-25 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The crystal structure of the Calystegia sepium agglutinin reveals a novel quaternary arrangement of lectin subunits with a beta-prism fold
J.Biol.Chem., 279, 2004
|
|
