7JJF
 
 | | Sarcin-ricin loop with modified residue. | | Descriptor: | MAGNESIUM ION, RNA/DNA (27-mer) | | Authors: | Harp, J.M, Pallan, P.S, Egli, M. | | Deposit date: | 2020-07-25 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Incorporating a Thiophosphate Modification into a Common RNA Tetraloop Motif Causes an Unanticipated Stability Boost.
Biochemistry, 59, 2020
|
|
7JK4
 
 | | Structure of Drosophila ORC bound to AT-rich DNA and Cdc6 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein, DNA (34-MER), ... | | Authors: | Schmidt, J.M, Bleichert, F. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6.
Nat Commun, 11, 2020
|
|
7JGS
 
 | |
1DE3
 
 | | SOLUTION STRUCTURE OF THE CYTOTOXIC RIBONUCLEASE ALPHA-SARCIN | | Descriptor: | RIBONUCLEASE ALPHA-SARCIN | | Authors: | Perez-Canadillas, J.M, Campos-Olivas, R, Santoro, J, Lacadena, J, Martinez del Pozo, A, Gavilanes, J.G, Rico, M, Bruix, M. | | Deposit date: | 1999-11-12 | | Release date: | 2000-06-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The highly refined solution structure of the cytotoxic ribonuclease alpha-sarcin reveals the structural requirements for substrate recognition and ribonucleolytic activity.
J.Mol.Biol., 299, 2000
|
|
1DIR
 
 | | CRYSTAL STRUCTURE OF A MONOCLINIC FORM OF DIHYDROPTERIDINE REDUCTASE FROM RAT LIVER | | Descriptor: | DIHYDROPTERIDINE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Varughese, K.I, Su, Y, Skinner, M.M, Matthews, D.A, Whitely, J.M, Xuong, N.H. | | Deposit date: | 1994-04-18 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a monoclinic form of dihydropteridine reductase from rat liver.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1GGI
 
 | | CRYSTAL STRUCTURE OF AN HIV-1 NEUTRALIZING ANTIBODY 50.1 IN COMPLEX WITH ITS V3 LOOP PEPTIDE ANTIGEN | | Descriptor: | HIV-1 V3 LOOP PEPTIDE ANTIGEN, IGG2A 50.1 FAB (HEAVY CHAIN), IGG2A 50.1 FAB (LIGHT CHAIN) | | Authors: | Stanfield, R.L, Rini, J.M, Wilson, I.A. | | Deposit date: | 1993-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
4MJ3
 
 | | Haloalkane dehalogenase DmrA from Mycobacterium rhodesiae JS60 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase, POTASSIUM ION | | Authors: | Fung, H, Gadd, M.S, Guss, J.M, Matthews, J.M. | | Deposit date: | 2013-09-03 | | Release date: | 2015-02-25 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and biophysical characterisation of haloalkane dehalogenases DmrA and DmrB in Mycobacterium strain JS60 and their role in growth on haloalkanes.
Mol.Microbiol., 97, 2015
|
|
8FHL
 
 | |
8FHU
 
 | |
4PCY
 
 | |
1TVK
 
 | | The binding mode of epothilone A on a,b-tubulin by electron crystallography | | Descriptor: | EPOTHILONE A, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nettles, J.H, Li, H, Cornett, B, Krahn, J.M, Snyder, J.P, Downing, K.H. | | Deposit date: | 2004-06-29 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.89 Å) | | Cite: | The binding mode of epothilone A on alpha,beta-tubulin by electron crystallography
Science, 305, 2004
|
|
1CBI
 
 | | APO-CELLULAR RETINOIC ACID BINDING PROTEIN I | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN I | | Authors: | Thompson, J.R, Bratt, J.M, Banaszak, L.J. | | Deposit date: | 1995-07-12 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of cellular retinoic acid binding protein I shows increased access to the binding cavity due to formation of an intermolecular beta-sheet.
J.Mol.Biol., 252, 1995
|
|
1U68
 
 | | DHNA 7,8 DIHYDRONEOPTERIN COMPLEX | | Descriptor: | 2-AMINO-7,8-DIHYDRO-6-(1,2,3-TRIHYDROXYPROPYL)-4(1H)-PTERIDINONE, Dihydroneopterin aldolase | | Authors: | Sanders, W.J, Nienaber, V.L, Lerner, C.G, McCall, J.O, Merrick, S.M, Swanson, S.J, Harlan, J.E, Stoll, V.S, Stamper, G.F, Betz, S.F, Condroski, K.R, Meadows, R.P, Severin, J.M, Walter, K.A, Magdalinos, P, Jakob, C.G, Wagner, R, Beutel, B.A. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent inhibitors of dihydroneopterin aldolase using CrystaLEAD high-throughput X-ray crystallographic screening and structure-directed lead optimization.
J.MED.CHEM., 47, 2004
|
|
1UVM
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 5NT RNA conformation A | | Descriptor: | 5'-R(*UP*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
4MWI
 
 | | Crystal structure of the human MLKL pseudokinase domain | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, GLYCEROL, Mixed lineage kinase domain-like protein | | Authors: | Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2013-09-25 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the evolution of divergent nucleotide-binding mechanisms among pseudokinases revealed by crystal structures of human and mouse MLKL.
Biochem.J., 457, 2014
|
|
1UO4
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4, iodobenzene | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UCI
 
 | | Mutants of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Takano, K, Scholtz, J.M, Sacchettini, J.C, Pace, C.N. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The contribution of polar group burial to protein stability is strongly context-dependent
J.Biol.Chem., 278, 2003
|
|
1UZI
 
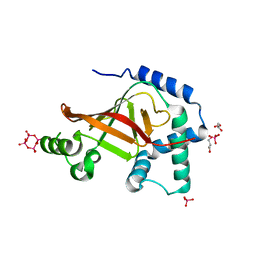 | | C3 EXOENZYME FROM CLOSTRIDIUM BOTULINUM, TETRAGONAL FORM | | Descriptor: | CYCLO-TETRAMETAVANADATE, GLYCEROL, MONO-ADP-RIBOSYLTRANSFERASE C3, ... | | Authors: | Evans, H.R, Holloway, D.E, Sutton, J.M, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2004-03-12 | | Release date: | 2004-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | C3 Exoenzyme from Clostridium Botulinum: Structure of a Tetragonal Crystal Form and a Reassessment of Nad-Induced Flexure
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1BRD
 
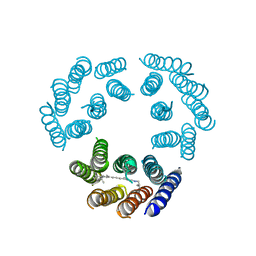 | | Model for the structure of Bacteriorhodopsin based on high-resolution Electron Cryo-microscopy | | Descriptor: | BACTERIORHODOPSIN PRECURSOR, RETINAL | | Authors: | Henderson, R, Baldwin, J.M, Ceska, T.A, Zemlin, F, Beckmann, E, Downing, K.H. | | Deposit date: | 1990-05-23 | | Release date: | 1991-04-15 | | Last modified: | 2024-04-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy.
J.Mol.Biol., 213, 1990
|
|
1WO3
 
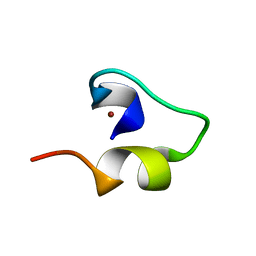 | | Solution structure of Minimal Mutant 1 (MM1): Multiple alanine mutant of non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
2YOI
 
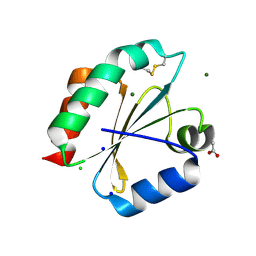 | | Crystal Structure of Ancestral Thioredoxin Relative to Last Eukaryotes Common Ancestor (LECA) from the Precambrian Period | | Descriptor: | ACETATE ION, CHLORIDE ION, LECA THIOREDOXIN, ... | | Authors: | Gavira, J.A, Ingles-Prieto, A, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-10-24 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conservation of protein structure over four billion years.
Structure, 21, 2013
|
|
2YPM
 
 | |
1TYF
 
 | |
1WO6
 
 | | Solution structure of Designed Functional Finger 5 (DFF5): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WPD
 
 | | Evidence for domain-specific recognition of SK and Kv channels by MTX and HsTx1 scorpion toxins | | Descriptor: | Potassium channel toxin alpha-KTx 6.2,Potassium channel toxin alpha-KTx 6.3 | | Authors: | Regaya, I, Beeton, C, Ferrat, G, Andreotti, N, Chandy, G.K, Darbon, H, De Waard, M, Sabatier, J.M. | | Deposit date: | 2004-09-01 | | Release date: | 2004-10-19 | | Last modified: | 2019-12-18 | | Method: | SOLUTION NMR | | Cite: | Evidence for domain-specific recognition of SK and Kv channels by MTX and HsTx1 scorpion toxins
J.Biol.Chem., 279, 2004
|
|
