7B7E
 
 | | Room temperature X-ray structure of perdeuterated PLL lectin | | Descriptor: | PLL lectin | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
4WSS
 
 | | The crystal structure of hemagglutinin form A/chicken/New York/14677-13/1998 in complex with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
4WSV
 
 | | The crystal structure of hemagglutinin from A/Taiwan/1/2013 in complex with 6'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
7B7C
 
 | | Room temperature X-ray structure of perdeuterated PLL lectin in complex with L-fucose | | Descriptor: | PLL lectin, alpha-L-fucopyranose, beta-L-fucopyranose | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
7PRG
 
 | | Joint X-ray/neutron room temperature structure of perdeuterated LecB lectin in complex with perdeuterated fucose | | Descriptor: | CALCIUM ION, Fucose-binding lectin, SULFATE ION, ... | | Authors: | Gajdos, L, Blakeley, M.P, Haertlein, M, Forsyth, T.V, Devos, J.M, Imberty, A. | | Deposit date: | 2021-09-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography reveals mechanisms used by Pseudomonas aeruginosa for host-cell binding.
Nat Commun, 13, 2022
|
|
7PSY
 
 | | X-ray crystal structure of perdeuterated LecB lectin in complex with perdeuterated fucose | | Descriptor: | CALCIUM ION, Fucose-binding lectin, SULFATE ION, ... | | Authors: | Gajdos, L, Blakeley, M.P, Haertlein, M, Forsyth, T.V, Devos, J.M, Imberty, A. | | Deposit date: | 2021-09-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Neutron crystallography reveals mechanisms used by Pseudomonas aeruginosa for host-cell binding.
Nat Commun, 13, 2022
|
|
7ZAN
 
 | |
7B1P
 
 | | Crystal Structure of Human BACE-1 in Complex with Compound 38a (NB-854) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(3~{R},6~{R})-5-azanyl-3,6-dimethyl-6-(trifluoromethyl)-2~{H}-1,4-oxazin-3-yl]phenyl]-5-bromanyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-25 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
7YX5
 
 | | Structure of the Mimivirus genomic fibre in its relaxed 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7B1Q
 
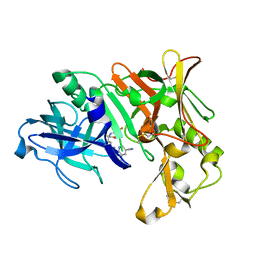 | | Crystal Structure of Human BACE-1 in Complex with Compound NB-360 (compound 54) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(3~{R},6~{R})-5-azanyl-3,6-dimethyl-6-(trifluoromethyl)-2~{H}-1,4-oxazin-3-yl]-4-fluoranyl-phenyl]-5-cyano-3-methyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-25 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
7S15
 
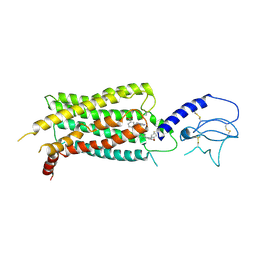 | | GLP-1 receptor bound with Pfizer small molecule agonist | | Descriptor: | 2-[(4-{6-[(2,4-difluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-[(1-ethyl-1H-imidazol-5-yl)methyl]-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor | | Authors: | Liu, Y, Dias, J.M, Han, S. | | Deposit date: | 2021-09-01 | | Release date: | 2022-06-08 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Small-Molecule Oral Agonist of the Human Glucagon-like Peptide-1 Receptor.
J.Med.Chem., 65, 2022
|
|
7YX4
 
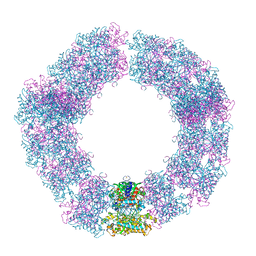 | | Structure of the Mimivirus genomic fibre in its compact 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7B1E
 
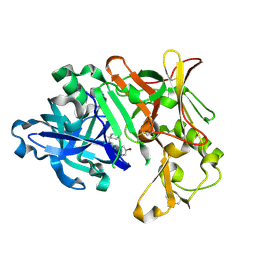 | | BACE1 IN COMPLEX WITH compound 3 (NB-641) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(4~{S})-2-azanyl-4-methyl-5,6-dihydro-1,3-thiazin-4-yl]phenyl]-5-bromanyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
7YX3
 
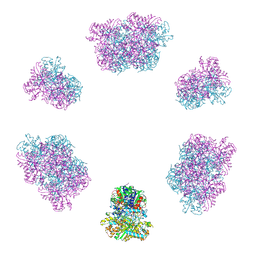 | | Structure of the Mimivirus genomic fibre in its compact 6-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield.
Elife, 11, 2022
|
|
4WTW
 
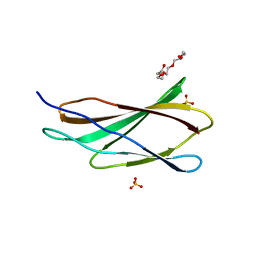 | | Crystal structure of the third FnIII domain of integrin beta4 | | Descriptor: | 1,2-ETHANEDIOL, Integrin beta-4, PENTAETHYLENE GLYCOL, ... | | Authors: | Alonso-Garcia, N, Urien, H, de Pereda, J.M. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Combination of X-ray crystallography, SAXS and DEER to obtain the structure of the FnIII-3,4 domains of integrin alpha6beta4
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7Q0L
 
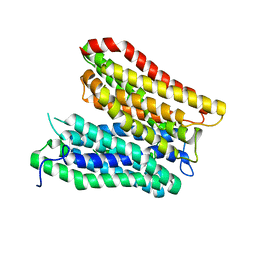 | | Crystal structure of the peptide transporter YePEPT-K314A at 2.93 A | | Descriptor: | Peptide transporter YePEPT | | Authors: | Jeckelmann, J.M, Stauffer, M, Ilgue, H, Boggavarapu, R, Fotiadis, D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Peptide transporter structure reveals binding and action mechanism of a potent PEPT1 and PEPT2 inhibitor.
Commun Chem, 5, 2022
|
|
7Q0M
 
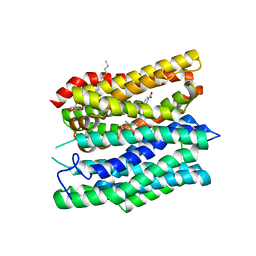 | | Crystal structure of the peptide transporter YePEPT-K314A in complex with LZNV at 2.66 A | | Descriptor: | (2~{S})-2-[[(2~{S})-2-azanyl-6-[(4-nitrophenyl)methoxycarbonylamino]hexanoyl]amino]-3-methyl-butanoic acid, Peptide transporter YePEPT, UNDECYL-MALTOSIDE | | Authors: | Jeckelmann, J.M, Stauffer, M, Ilgue, H, Fotiadis, D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Peptide transporter structure reveals binding and action mechanism of a potent PEPT1 and PEPT2 inhibitor.
Commun Chem, 5, 2022
|
|
7PON
 
 | |
7Z3W
 
 | | Crystal structure of the AAL160 Fab | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, AAL160 Fab heavy-chain, AAL160 Fab light-chain | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2022-03-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | "Redirecting an anti-IL-1 beta antibody to bind a new, unrelated and computationally predicted epitope on hIL-17A".
Commun Biol, 6, 2023
|
|
7Z4T
 
 | | AAL160 FAB IN COMPLEX WITH HUMAN INTERLEUKIN-1 BETA | | Descriptor: | AAL160 Fab heavy-chain, AAL160 light-chain, Interleukin-1 beta | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2022-03-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | "Redirecting an anti-IL-1 beta antibody to bind a new, unrelated and computationally predicted epitope on hIL-17A".
Commun Biol, 6, 2023
|
|
7Z2M
 
 | |
7APW
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-(benzo[d]thiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-(benzo[d]thiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, A.M, Kolos, J.M, Pomplun, S, Riess, B, Purder, P, Merz, S, Bracher, A, Meyners, C, Krewald, V, Hausch, F. | | Deposit date: | 2020-10-20 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Picomolar FKBP inhibitors enabled by a single water-displacing methyl group in bicyclic [4.3.1] aza-amides.
Chem Sci, 12, 2021
|
|
8OXJ
 
 | |
4X08
 
 | | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms. | | Descriptor: | Eosinophil cationic protein, SULFATE ION | | Authors: | Blanco, J.A, Garcia, J.M, Salazar, V.A, Sanchez, D, Moussauoi, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms.
To Be Published
|
|
4WST
 
 | | The crystal structure of hemagglutinin from A/Taiwan/1/2013 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Yang, H, Carney, P.J, Chang, J, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
