3J48
 
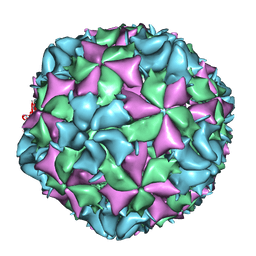 | | Cryo-EM structure of Poliovirus 135S particles | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Butan, C, Fiman, D.J, Hogle, J.M. | | Deposit date: | 2013-06-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-Electron Microscopy Reconstruction Shows Poliovirus 135S Particles Poised for Membrane Interaction and RNA Release.
J.Virol., 88, 2014
|
|
2XGF
 
 | | Structure of the bacteriophage T4 long tail fibre needle-shaped receptor-binding tip | | Descriptor: | CARBONATE ION, FE (II) ION, LONG TAIL FIBER PROTEIN P37 | | Authors: | Bartual, S.G, Otero, J.M, Garcia-Doval, C, Llamas-Saiz, A.L, Kahn, R, Fox, G.C, van Raaij, M.J. | | Deposit date: | 2010-06-03 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the bacteriophage T4 long tail fiber receptor-binding tip.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
3J4B
 
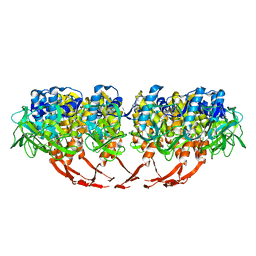 | | Structure of T7 gatekeeper protein (gp11) | | Descriptor: | Tail tubular protein A | | Authors: | Cuervo, A, Pulido-Cid, M, Chagoyen, M, Arranz, R, Gonzalez-Garcia, V.A, Garcia-Doval, C, Caston, J.R, Valpuesta, J.M, van Raaij, M.J, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2013-07-09 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural characterization of the bacteriophage t7 tail machinery.
J.Biol.Chem., 288, 2013
|
|
3K4I
 
 | | CRYSTAL STRUCTURE OF uncharacterized protein PSPTO_3204 from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, uncharacterized protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | CRYSTAL STRUCTURE OF uncharacterized protein PSPTO_3204 from Pseudomonas syringae pv. tomato
str. DC3000
To be Published
|
|
2BOV
 
 | | Molecular recognition of an ADP-ribosylating Clostridium botulinum C3 exoenzyme by RalA GTPase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MONO-ADP-RIBOSYLTRANSFERASE C3, ... | | Authors: | Holbourn, K.P, Sutton, J.M, Evans, H.R, Shone, C.C, Acharya, K.R. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Molecular Recognition of an Adp-Ribosylating Clostridium Botulinum C3 Exoenzyme by Rala Gtpase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3K5T
 
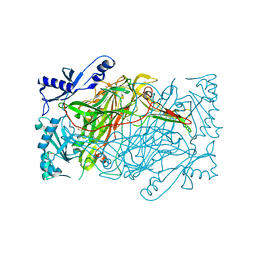 | | Crystal structure of human diamine oxidase in space group C2221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | McGrath, A.P, Guss, J.M. | | Deposit date: | 2009-10-08 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | A new crystal form of human diamine oxidase.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3KAJ
 
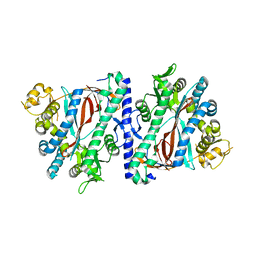 | | Apoenzyme structure of homoglutathione synthetase from Glycine max in open conformation | | Descriptor: | Homoglutathione synthetase | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
3J8F
 
 | | Cryo-EM reconstruction of poliovirus-receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, ... | | Authors: | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
1GUW
 
 | | STRUCTURE OF THE CHROMODOMAIN FROM MOUSE HP1beta IN COMPLEX WITH THE LYSINE 9-METHYL HISTONE H3 N-TERMINAL PEPTIDE, NMR, 25 STRUCTURES | | Descriptor: | CHROMOBOX PROTEIN HOMOLOG 1, HISTONE H3.1 | | Authors: | Nielsen, P.R, Nietlispach, D, Mott, H.R, Callaghan, J.M, Bannister, A, Kouzarides, T, Murzin, A.G, Murzina, N.V, Laue, E.D. | | Deposit date: | 2002-02-01 | | Release date: | 2002-03-12 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | Structure of the Hp1 Chromodomain Bound to Histone H3 Methylated at Lysine 9
Nature, 416, 2002
|
|
2P2U
 
 | | Crystal structure of putative host-nuclease inhibitor protein Gam from Desulfovibrio vulgaris | | Descriptor: | Host-nuclease inhibitor protein Gam, putative | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Zhang, F, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of putative host-nuclease inhibitor protein Gam from Desulfovibrio vulgaris
To be Published
|
|
1YRM
 
 | |
1RZ6
 
 | | Di-haem Cytochrome c Peroxidase, Form IN | | Descriptor: | CITRIC ACID, Cytochrome c peroxidase, HEME C | | Authors: | Dias, J.M, Alves, T, Bonifacio, C, Pereira, A, Bourgeois, D, Moura, I, Romao, M.J. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the mechanism of Ca(2+) activation of the di-heme cytochrome c peroxidase from Pseudomonas nautica 617.
Structure, 12, 2004
|
|
1YSN
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-X | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
3K2W
 
 | | CRYSTAL STRUCTURE OF betaine-aldehyde dehydrogenase FROM Pseudoalteromonas atlantica T6c | | Descriptor: | ACETATE ION, Betaine-aldehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF betaine-aldehyde dehydrogenase FROM Pseudoalteromonas atlantica
To be Published
|
|
3K4U
 
 | | CRYSTAL STRUCTURE OF putative binding component of ABC transporter from Wolinella succinogenes DSM 1740 complexed with lysine | | Descriptor: | BINDING COMPONENT OF ABC TRANSPORTER, LYSINE | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-06 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | CRYSTAL STRUCTURE OF putative binding component of ABC transporter from Wolinella succinogenes
DSM 1740 complexed with lysine
To be Published
|
|
2P4U
 
 | | Crystal structure of acid phosphatase 1 (Acp1) from Mus musculus | | Descriptor: | Acid phosphatase 1, PHOSPHATE ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Xu, W, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-13 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2XSM
 
 | | Crystal structure of the mammalian cytosolic chaperonin CCT in complex with tubulin | | Descriptor: | CCT | | Authors: | Munoz, I.G, Yebenes, H, Zhou, M, Mesa, P, Serna, M, Bragado-Nilsson, E, Beloso, A, Robinson, C.V, Valpuesta, J.M, Montoya, G. | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal Structure of the Open Conformation of the Mammalian Chaperonin Cct in Complex with Tubulin.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3J9F
 
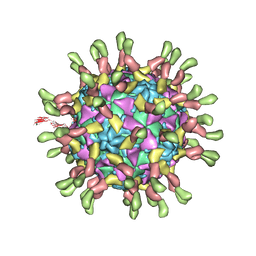 | | Poliovirus complexed with soluble, deglycosylated poliovirus receptor (Pvr) at 4 degrees C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-02-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
1HA4
 
 | |
2OZA
 
 | | Structure of p38alpha complex | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14 | | Authors: | White, A, Pargellis, C.A, Studts, J.M, Werneburg, B.G, Farmer II, B.T. | | Deposit date: | 2007-02-25 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of MAPK-activated protein kinase 2:p38 assembly
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2P27
 
 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with Mg2+ at 1.9 A resolution | | Descriptor: | MAGNESIUM ION, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
3JW3
 
 | |
3JWM
 
 | | Crystal structure of Bacillus anthracis dihydrofolate reductase complexed with NADPH and (S)-2,4-diamino-5-(3-methoxy-3-(3,4,5-trimethoxyphenyl)prop-1-ynyl)-6-methylpyrimidine (UCP114A) | | Descriptor: | 5-[(3S)-3-methoxy-3-(3,4,5-trimethoxyphenyl)prop-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Anderson, A.C, Beierlein, J.M, Karri, N.G. | | Deposit date: | 2009-09-18 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Mutational Studies into Trimethoprim Resistance in Bacillus anthracis Dihydrofolate Reductase
To be Published
|
|
2Z25
 
 | | Thr110Val dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
2P5E
 
 | | Crystal Structures of High Affinity Human T-Cell Receptors Bound to pMHC Reveal Native Diagonal Binding Geometry | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, Cancer/testis antigen 1B, ... | | Authors: | Sami, M, Rizkallah, P.J, Dunn, S, Li, Y, Moysey, R, Vuidepot, A, Baston, E, Todorov, P, Molloy, P, Gao, F, Boulter, J.M, Jakobsen, B.K. | | Deposit date: | 2007-03-15 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of high affinity human T-cell receptors bound to peptide major
histocompatibility complex reveal native diagonal binding geometry
Protein Eng.Des.Sel., 20, 2007
|
|
