3FV9
 
 | | Crystal structure of putative mandelate racemase/muconatelactonizing enzyme from ROSEOVARIUS NUBINHIBENS ISM complexed with magnesium | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Malashkevich, V.N, Rutter, M, Bain, K.T, Lau, C, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-15 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative mandelate racemase/muconatelactonizing enzyme from ROSEOVARIUS NUBINHIBENS ISM complexed with magnesium
to be published
|
|
3B40
 
 | | Crystal structure of the probable dipeptidase PvdM from Pseudomonas aeruginosa | | Descriptor: | CADMIUM ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Dickey, M, Bain, K.T, Mendoza, M, Fong, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-23 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the probable dipeptidase PvdM from Pseudomonas aeruginosa.
To be Published
|
|
3LJI
 
 | | CRYSTAL STRUCTURE OF putative geranyltranstransferase from Pseudomonas fluorescens Pf-5 | | Descriptor: | Geranyltranstransferase | | Authors: | Malashkevich, V.N, Toro, R, Patskovsky, Y, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-26 | | Release date: | 2010-02-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | CRYSTAL STRUCTURE OF putative geranyltranstransferase from Pseudomonas fluorescens Pf-5
To be Published
|
|
3L7C
 
 | | Crystal Structure of Glycogen Phosphorylase DK4 complex | | Descriptor: | 1-(3-deoxy-3-fluoro-beta-D-glucopyranosyl)-5-fluoropyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Tsirkone, V.G, Lamprakis, C, Hayes, J.M, Skamnaki, V, Drakou, C, Zographos, S.E, Leonidas, D.D. | | Deposit date: | 2009-12-28 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | 1-(3-Deoxy-3-fluoro-beta-d-glucopyranosyl) pyrimidine derivatives as inhibitors of glycogen phosphorylase b: Kinetic, crystallographic and modelling studies.
Bioorg.Med.Chem., 18, 2010
|
|
1L5B
 
 | | DOMAIN-SWAPPED CYANOVIRIN-N DIMER | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, SODIUM ION, cyanovirin-N | | Authors: | Barrientos, L.G, Louis, J.M, Botos, I, Mori, T, Han, Z, O'Keefe, B.R, Boyd, M.R, Wlodawer, A, Gronenborn, A.M. | | Deposit date: | 2002-03-06 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures.
Structure, 10, 2002
|
|
3BCS
 
 | | Glycogen Phosphorylase complex with 1(-D-glucopyranosyl) uracil | | Descriptor: | 1-beta-D-glucopyranosylpyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Sovantzis, D.A, Hadjiloi, T, Hayes, J.M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G. | | Deposit date: | 2007-11-13 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D-Glucopyranosyl pyrimidine nucleoside binding to muscle glycogen phosphorylase b
To be Published
|
|
3FV7
 
 | | OXA-24 beta-lactamase complex with SA4-44 inhibitor | | Descriptor: | (2S)-2-[[2-methanoyl-7-(methoxycarbonylamino)indolizin-3-yl]amino]-3-methyl-3-sulfino-butanoic acid, Beta-lactamase OXA-24 | | Authors: | Bou, G, Santillana, E, Sheri, A, Beceiro, A, Sampson, J.M, Kalp, M, Bethel, C.R, Distler, A.M, Drawz, S.M, Pagadala, S.R, Van den Akker, F, Bonomo, R.A, Romero, A, Buynak, J.D. | | Deposit date: | 2009-01-15 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase.
J.Am.Chem.Soc., 132, 2010
|
|
3P6H
 
 | |
3FRN
 
 | | CRYSTAL STRUCTURE OF flagellar protein FlgA FROM Thermotoga maritima MSB8 | | Descriptor: | Flagellar protein FlgA, GLYCEROL | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Hu, S, Bain, K, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CRYSTAL STRUCTURE OF flagellar protein FlgA FROM Thermotoga maritima
To be Published
|
|
3LIM
 
 | | Crystal structure of the pore forming toxin frac from sea anemone actinia fragacea | | Descriptor: | Fragaceatoxin C, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Mechaly, A.E, Bellomio, A, Morante, K, Gonzalez-Manas, J.M, Guerin, D.M.A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the oligomerization and architecture of eukaryotic membrane pore-forming toxins.
Structure, 19, 2011
|
|
4A6W
 
 | | X-ray structures of oxazole hydroxamate EcMetAp-Mn complexes | | Descriptor: | 5-(2-chlorophenyl)-N-hydroxy-1,3-oxazole-2-carboxamide, MANGANESE (II) ION, METHIONINE AMINOPEPTIDASE | | Authors: | Huguet, F, Melet, A, AlvesdeSousa, R, Lieutaud, A, Chevalier, J, Deschamps, P, Tomas, A, Leulliot, N, Pages, J.M, Artaud, I. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Hydroxamic Acids as Potent Inhibitors of Fe(II) and Mn(II) E. Coli Methionine Aminopeptidase: Biological Activities and X-Ray Structures of Oxazole Hydroxamate-Ecmetap-Mn Complexes.
Chemmedchem, 7, 2012
|
|
3B1W
 
 | | Crystal structure of an S. thermophilus NFeoB E67A mutant bound to GDP | | Descriptor: | Ferrous iron uptake transporter protein B, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-07-15 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A suite of Switch I and Switch II mutant structures from the G-protein domain of FeoB
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3LKE
 
 | | Crystal structure of enoyl-CoA hydratase from Bacillus halodurans | | Descriptor: | Enoyl-CoA hydratase, GLYCEROL | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of enoyl-CoA hydratase from Bacillus halodurans
To be Published
|
|
3L60
 
 | | Crystal structure of branched-chain alpha-keto acid dehydrogenase subunit e2 from mycobacterium tuberculosis | | Descriptor: | BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE, UNKNOWN LIGAND | | Authors: | Zencheck, W.D, Bonanno, J.B, Patskovsky, Y, Toro, R, Freeman, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Branched-Chain Alpha-Keto Acid Dehydrogenase Subunit E2 from Mycobacterium Tuberculosis
To be Published
|
|
2F84
 
 | |
3FSO
 
 | |
1RJO
 
 | | AGAO + Xe | | Descriptor: | COPPER (II) ION, GLYCEROL, Phenylethylamine oxidase, ... | | Authors: | Guss, J.M, Trambaiolo, D.M, Duff, A.P. | | Deposit date: | 2003-11-19 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Using Xenon as a Probe for Dioxygen-binding Sites in Copper Amine Oxidases
J.Mol.Biol., 344, 2004
|
|
3B9J
 
 | | Structure of Xanthine Oxidase with 2-hydroxy-6-methylpurine | | Descriptor: | 6-methyl-3,9-dihydro-2H-purin-2-one, CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Pauff, J.M, Zhang, J, Bell, C.E, Hille, R. | | Deposit date: | 2007-11-05 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate orientation in xanthine oxidase: crystal structure of enzyme in reaction with 2-hydroxy-6-methylpurine.
J.Biol.Chem., 283, 2008
|
|
3L8D
 
 | | Crystal structure of methyltransferase from Bacillus Thuringiensis | | Descriptor: | Methyltransferase, POTASSIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of methyltransferase from Bacillus Thuringiensis
To be Published
|
|
3L8K
 
 | | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dihydrolipoyl dehydrogenase, PHOSPHATE ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus
To be Published
|
|
3PS4
 
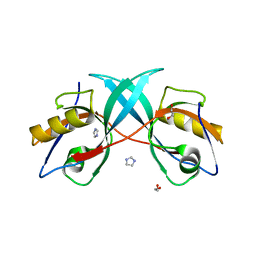 | | PDZ domain from Human microtubule-associated serine/threonine-protein kinase 1 | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Microtubule-associated serine/threonine-protein kinase 1 | | Authors: | Ugochukwu, E, Wang, J, Krojer, T, Muniz, J.R.C, Sethi, R, Pike, A.C.W, Roos, A, Salah, E, Cocking, R, Savitsky, P, Doyle, D.A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-30 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PDZ domain from Human microtubule-associated serine/threonine-protein kinase 1
TO BE PUBLISHED
|
|
2GCZ
 
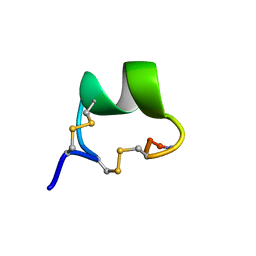 | | Solution Structure of alpha-Conotoxin OmIA | | Descriptor: | Alpha-conotoxin OmIA | | Authors: | Chi, S.-W, Kim, D.-H, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | Deposit date: | 2006-03-15 | | Release date: | 2006-07-25 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of a neuronal nicotinic acetylcholine receptor antagonist alpha-conotoxin OmIA that discriminates alpha3 vs. alpha6 nAChR subtypes
Biochem.Biophys.Res.Commun., 345, 2006
|
|
3BCU
 
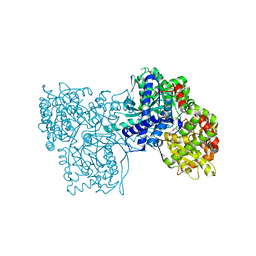 | | Glucogen Phosphorylase complex with thymidine | | Descriptor: | Glycogen phosphorylase, muscle form, THYMIDINE | | Authors: | Sovantzis, D.A, Hadjiloi, T, Hayes, J.M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G. | | Deposit date: | 2007-11-13 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | D-Glucopyranosyl pyrimidine nucleoside binding to muscle glycogen phosphorylase b
To be Published
|
|
1L5I
 
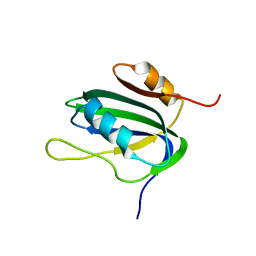 | | 30-CONFORMER NMR ENSEMBLE OF THE N-TERMINAL, DNA-BINDING DOMAIN OF THE REPLICATION INITIATION PROTEIN FROM A GEMINIVIRUS (TOMATO YELLOW LEAF CURL VIRUS-SARDINIA) | | Descriptor: | Rep protein | | Authors: | Campos-Olivas, R, Louis, J.M, Clerot, D, Gronenborn, B, Gronenborn, A.M. | | Deposit date: | 2002-03-07 | | Release date: | 2002-09-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of a replication initiator unites diverse aspects of nucleic acid metabolism
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1RDT
 
 | | Crystal Structure of a new rexinoid bound to the RXRalpha ligand binding doamin in the RXRalpha/PPARgamma heterodimer | | Descriptor: | (S)-(2E)-3[4-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-2-NAPHTHALENYL)TETRAHYDRO-1-BENZOFURAN-2-YL]-2-PROPENOIC ACID, 2-(2-BENZOYL-PHENYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, LxxLL motif coactivator, ... | | Authors: | Haffner, C.D, Lenhard, J.M, Miller, A.B, McDougald, D.L, Dwornik, K, Ittoop, O.R, Gampe Jr, R.T, Xu, H.E, Blanchard, S, Montana, V.G. | | Deposit date: | 2003-11-06 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of potent retinoid X receptor alpha agonists.
J.Med.Chem., 47, 2004
|
|
