4X93
 
 | | Crystal structure of Lysosomal Phospholipase A2 crystallized in the presence of methyl arachidonyl fluorophosphonate (tetragonal form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Glukhova, A, Tesmer, J.J.G. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of lysosomal phospholipase A2 and lecithin:cholesterol acyltransferase.
Nat Commun, 6, 2015
|
|
1BAP
 
 | |
4X92
 
 | |
1W4U
 
 | | NMR solution structure of the ubiquitin conjugating enzyme UbcH5B | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2-17 KDA 2 | | Authors: | Houben, K, Dominguez, C, Van Schaik, F.M.A, Timmers, H.T.M, Bonvin, A.M.J.J, Boelens, R. | | Deposit date: | 2004-07-29 | | Release date: | 2004-11-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ubiquitin-Conjugating Enzyme Ubch5B
J.Mol.Biol., 344, 2004
|
|
2M7R
 
 | | Nmda receptor antagonist, conantokin bk-b, nmr, 20 structure | | Descriptor: | CONANTOKIN BK-B | | Authors: | Curtice, K.J, Platt, R.J, Skalicky, J.J, Horvath, M.P, Twede, V.D. | | Deposit date: | 2013-04-29 | | Release date: | 2014-02-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | From molecular phylogeny towards differentiating pharmacology for NMDA receptor subtypes.
Toxicon, 81C, 2014
|
|
2MAG
 
 | | NMR STRUCTURE OF MAGAININ 2 IN DPC MICELLES, 10 STRUCTURES | | Descriptor: | MAGAININ 2 | | Authors: | Gesell, J.J, Zasloff, M, Opella, S.J. | | Deposit date: | 1997-12-19 | | Release date: | 1998-04-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Two-dimensional 1H NMR experiments show that the 23-residue magainin antibiotic peptide is an alpha-helix in dodecylphosphocholine micelles, sodium dodecylsulfate micelles, and trifluoroethanol/water solution.
J.Biomol.NMR, 9, 1997
|
|
4X96
 
 | |
4X95
 
 | |
4X97
 
 | |
1W8X
 
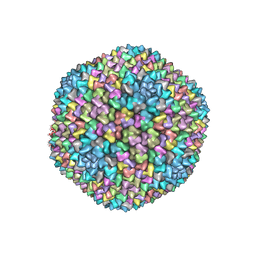 | | Structural analysis of PRD1 | | Descriptor: | MAJOR CAPSID PROTEIN (PROTEIN P3), PROTEIN P16, PROTEIN P30, ... | | Authors: | Abrescia, N.G.A, Cockburn, J.J.B, Grimes, J.M, Sutton, G.C, Diprose, J.M, Butcher, S.J, Fuller, S.D, San Martin, C, Burnett, R.M, Stuart, D.I, Bamford, D.H, Bamford, J.K.H. | | Deposit date: | 2004-10-01 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Insights Into Assembly from Structural Analysis of Bacteriophage Prd1.
Nature, 432, 2004
|
|
1AZS
 
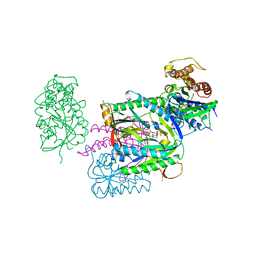 | |
1ARR
 
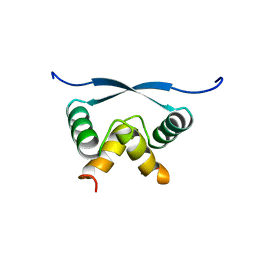 | | RELAXATION MATRIX REFINEMENT OF THE SOLUTION STRUCTURE OF THE ARC REPRESSOR | | Descriptor: | ARC REPRESSOR | | Authors: | Bonvin, A.M.J.J, Vis, H, Burgering, M.J.M, Breg, J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-08-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the Arc repressor using relaxation matrix calculations.
J.Mol.Biol., 236, 1994
|
|
1ARQ
 
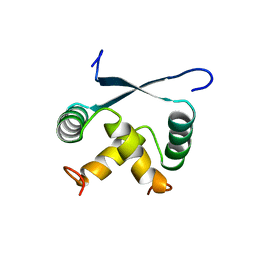 | | RELAXATION MATRIX REFINEMENT OF THE SOLUTION STRUCTURE OF THE ARC REPRESSOR | | Descriptor: | ARC REPRESSOR | | Authors: | Bonvin, A.M.J.J, Vis, H, Burgering, M.J.M, Breg, J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-08-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the Arc repressor using relaxation matrix calculations.
J.Mol.Biol., 236, 1994
|
|
1YM7
 
 | | G Protein-Coupled Receptor Kinase 2 (GRK2) | | Descriptor: | Beta-adrenergic receptor kinase 1 | | Authors: | Lodowski, D.T, Barnhill, J.F, Pyskadlo, R.M, Ghirlando, R, Sterne-Marr, R, Tesmer, J.J.G. | | Deposit date: | 2005-01-20 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The role of Gbetagamma and domain interfaces in the activation of G protein-coupled receptor kinase 2
Biochemistry, 44, 2005
|
|
1VLB
 
 | | STRUCTURE REFINEMENT OF THE ALDEHYDE OXIDOREDUCTASE FROM DESULFOVIBRIO GIGAS AT 1.28 A | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, CHLORIDE ION, ... | | Authors: | Rebelo, J.M, Dias, J.M, Huber, R, Moura, J.J.G, Romao, M.J. | | Deposit date: | 2004-07-20 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure refinement of the aldehyde oxidoreductase from Desulfovibrio gigas (MOP) at 1.28 A
J.Biol.Inorg.Chem., 6, 2001
|
|
2NIP
 
 | | NITROGENASE IRON PROTEIN FROM AZOTOBACTER VINELANDII | | Descriptor: | IRON/SULFUR CLUSTER, NITROGENASE IRON PROTEIN | | Authors: | Komiya, H, Georgiadis, M.M, Chakrabarti, P, Woo, D, Kornuc, J.J, Rees, D.C. | | Deposit date: | 1998-05-11 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational variability in structures of the nitrogenase iron proteins from Azotobacter vinelandii and Clostridium pasteurianum.
J.Mol.Biol., 280, 1998
|
|
1ZMT
 
 | | Structure of haloalcohol dehalogenase HheC of Agrobacterium radiobacter AD1 in complex with (R)-para-nitro styrene oxide, with a water molecule in the halide-binding site | | Descriptor: | (R)-PARA-NITROSTYRENE OXIDE, Haloalcohol dehalogenase HheC | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Villa, A, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-05-10 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Enantioselectivity of an Epoxide Ring Opening Reaction Catalyzed by Halo Alcohol Dehalogenase HheC.
J.Am.Chem.Soc., 127, 2005
|
|
1XRK
 
 | | Crystal structure of a mutant bleomycin binding protein from Streptoalloteichus hindustanus displaying increased thermostability | | Descriptor: | BLEOMYCIN A2, Bleomycin resistance protein, SULFATE ION | | Authors: | Brouns, S.J.J, Wu, H, Akerboom, J, Turnbull, A.P, de Vos, W.M, Van der Oost, J. | | Deposit date: | 2004-10-15 | | Release date: | 2005-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering a selectable marker for hyperthermophiles
J.Biol.Chem., 280, 2005
|
|
1YY8
 
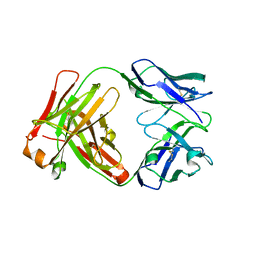 | | Crystal structure of the Fab fragment from the monoclonal antibody cetuximab/Erbitux/IMC-C225 | | Descriptor: | Cetuximab Fab Heavy chain, Cetuximab Fab Light chain | | Authors: | Li, S, Schmitz, K.R, Jeffrey, P.D, Wiltzius, J.J.W, Kussie, P, Ferguson, K.M. | | Deposit date: | 2005-02-24 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of the epidermal growth factor receptor by cetuximab
Cancer Cell, 7, 2005
|
|
1CCM
 
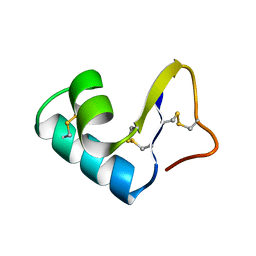 | | DIRECT NOE REFINEMENT OF CRAMBIN FROM 2D NMR DATA USING A SLOW-COOLING ANNEALING PROTOCOL | | Descriptor: | CRAMBIN | | Authors: | Bonvin, A.M.J.J, Rullmann, J.A.C, Lamerichs, R.M.J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-04-14 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | "Ensemble" iterative relaxation matrix approach: a new NMR refinement protocol applied to the solution structure of crambin.
Proteins, 15, 1993
|
|
1CCN
 
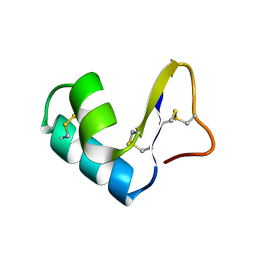 | | DIRECT NOE REFINEMENT OF CRAMBIN FROM 2D NMR DATA USING A SLOW-COOLING ANNEALING PROTOCOL | | Descriptor: | CRAMBIN | | Authors: | Bonvin, A.M.J.J, Rullmann, J.A.C, Lamerichs, R.M.J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-04-14 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Direct NOE refinement of biomolecular structures using 2D NMR data
J.Biomol.NMR, 1, 1991
|
|
1AZT
 
 | | GS-ALPHA COMPLEXED WITH GTP-GAMMA-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GS-ALPHA, MAGNESIUM ION, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1997-11-20 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the adenylyl cyclase activator Gsalpha
Science, 278, 1997
|
|
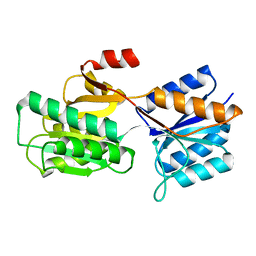
1APB
 
 | |
1ZCB
 
 | | Crystal structure of G alpha 13 in complex with GDP | | Descriptor: | G alpha i/13, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Nance, M.R, Tesmer, J.J.G. | | Deposit date: | 2005-04-11 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new approach to producing functional G alpha subunits yields the activated and deactivated structures of G alpha(12/13) proteins.
Biochemistry, 45, 2006
|
|
1AGR
 
 | | COMPLEX OF ALF4-ACTIVATED GI-ALPHA-1 WITH RGS4 | | Descriptor: | CITRIC ACID, GUANINE NUCLEOTIDE-BINDING PROTEIN G(I), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1997-03-25 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of RGS4 bound to AlF4--activated G(i alpha1): stabilization of the transition state for GTP hydrolysis.
Cell(Cambridge,Mass.), 89, 1997
|
|
